biopipen.ns.tcr
Tools to analyze single-cell TCR sequencing data
ImmunarchLoading(Proc) — Immuarch - Loading data</>ImmunarchFilter(Proc) — Immunarch - Filter data</>Immunarch(Proc) — Exploration of Single-cell and Bulk T-cell/Antibody Immune Repertoires</>SampleDiversity(Proc) — Sample diversity and rarefaction analysis</>CloneResidency(Proc) — Identification of clone residency</>Immunarch2VDJtools(Proc) — Convert immuarch format into VDJtools input formats.</>ImmunarchSplitIdents(Proc) — Split the data into multiple immunarch datasets by Idents from Seurat</>VJUsage(Proc) — Circos-style V-J usage plot displaying the frequency ofvarious V-J junctions using vdjtools. </>Attach2Seurat(Proc) — Attach the clonal information to a Seurat object as metadata</>CDR3Clustering(Proc) — Cluster the TCR/BCR clones by their CDR3 sequences</>TCRClusterStats(Proc) — Statistics of TCR clusters, generated byTCRClustering.</>CloneSizeQQPlot(Proc) — QQ plot of the clone sizes</>CDR3AAPhyschem(Proc) — CDR3 AA physicochemical feature analysis</>TESSA(Proc) — Tessa is a Bayesian model to integrate T cell receptor (TCR) sequenceprofiling with transcriptomes of T cells. </>TCRDock(Proc) — Using TCRDock to predict the structure of MHC-peptide-TCR complexes</>ScRepLoading(Proc) — Load the single cell TCR/BCR data into ascRepertoirecompatible object</>ScRepCombiningExpression(Proc) — Combine the scTCR/BCR data with the expression data</>ClonalStats(Proc) — Visualize the clonal information.</>
biopipen.ns.tcr.ImmunarchLoading(*args, **kwds) → Proc
Immuarch - Loading data
Load the raw data into immunarch object,
using immunarch::repLoad().
For the data path specified at TCRData in the input file, we will first find
filtered_contig_annotations.csv and filtered_config_annotations.csv.gz in the
path. If neighter of them exists, we will find all_contig_annotations.csv and
all_contig_annotations.csv.gz in the path and a warning will be raised
(You can find it at ./.pipen/<pipeline-name>/ImmunarchLoading/0/job.stderr).
If none of the files exists, an error will be raised.
This process will also generate a text file with the information for each cell.
The file will be saved at
./.pipen/<pipeline-name>/ImmunarchLoading/0/output/<prefix>.tcr.txt.
The file can be used by the SeuratMetadataMutater process to integrate the
TCR-seq data into the Seurat object for further integrative analysis.
envs.metacols can be used to specify the columns to be exported to the text file.
cache— Should we detect whether the jobs are cached?desc— The description of the process. Will use the summary fromthe docstring by default.dirsig— When checking the signature for caching, whether should we walkthrough the content of the directory? This is sometimes time-consuming if the directory is big.envs— The arguments that are job-independent, useful for common optionsacross jobs.envs_depth— How deep to update the envs when subclassed.error_strategy— How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
export— When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processesforks— How many jobs to run simultaneously?input— The keys for the input channelinput_data— The input data (will be computed for dependent processes)lang— The language for the script to run. Should be the path to theinterpreter iflangis not in$PATH.name— The name of the process. Will use the class name by default.nexts— Computed fromrequiresto build the process relationshipsnum_retries— How many times to retry to jobs once error occursorder— The execution order for this process. The bigger the numberis, the later the process will be executed. Default: 0. Note that the dependent processes will always be executed first. This doesn't work for start processes either, whose orders are determined byPipen.set_starts()output— The output keys for the output channel(the data will be computed)output_data— The output data (to pass to the next processes)plugin_opts— Options for process-level pluginsrequires— The dependency processesscheduler— The scheduler to run the jobsscheduler_opts— The options for the schedulerscript— The script template for the processsubmission_batch— How many jobs to be submited simultaneously.The program entrance for some schedulers may take too much resources when submitting a job or checking the job status. So we may use a smaller number here to limit the simultaneous submissions.template— Define the template engine to use.This could be either a template engine or a dict with keyengineindicating the template engine and the rest the arguments passed to the constructor of thepipen.template.Templateobject. The template engine could be either the name of the engine, currently jinja2 and liquidpy are supported, or a subclass ofpipen.template.Template. You can subclasspipen.template.Templateto use your own template engine.
metafile— The meta data of the samplesA tab-delimited file Two columns are required:- *
Sampleto specify the sample names. - *
TCRDatato assign the path of the data to the samples,
filtered_contig_annotations.csv, which doesn't have any name information.- *
metatxt— The meta data at cell level, which can be used to attach to the Seurat objectrdsfile— The RDS file with the data and metadata, which can be processed byotherimmunarchfunctions.
extracols(list) — The extra columns to be exported to the text file.You can refer to the immunarch documentation to get a sense for the full list of the columns. The columns may vary depending on the data source. The columns fromimmdata$metaand some core columns, includingBarcode,CDR3.aa,Clones,Proportion,V.name,J.name, andD.namewill be exported by default. You can use this option to specify the extra columns to be exported.mode— Either "single" for single chain data or "paired" forpaired chain data. Forsingle, only TRB chain will be kept atimmdata$data, information for other chains will be saved atimmdata$traandimmdata$multi.prefix— The prefix to the barcodes. You can use placeholder like{Sample}_to use the meta data from theimmunarchobject. The prefixed barcodes will be saved inout.metatxt. Theimmunarchobject keeps the original barcodes, but the prefix is saved atimmdata$prefix.
/// Note This option is useful because the barcodes for the cells from scRNA-seq data are usually prefixed with the sample name, for example,Sample1_AAACCTGAGAAGGCTA-1. However, the barcodes for the cells from scTCR-seq data are usually not prefixed with the sample name, for example,AAACCTGAGAAGGCTA-1. So we need to add the prefix to the barcodes for the scTCR-seq data, and it is easier for us to integrate the data from different sources later. ///tmpdir— The temporary directory to link all data files.Immunarchscans a directory to find the data files. If the data files are not in the same directory, we can link them to a temporary directory and pass the temporary directory toImmunarch. This option is useful when the data files are in different directories.
__init_subclass__()— Do the requirements inferring since we need them to build up theprocess relationship </>from_proc(proc,name,desc,envs,envs_depth,cache,export,error_strategy,num_retries,forks,input_data,order,plugin_opts,requires,scheduler,scheduler_opts,submission_batch)(Type) — Create a subclass of Proc using another Proc subclass or Proc itself</>gc()— GC process for the process to save memory after it's done</>log(level,msg,*args,logger)— Log message for the process</>run()— Init all other properties and jobs</>
pipen.proc.ProcMeta(name, bases, namespace, **kwargs)
Meta class for Proc
__call__(cls,*args,**kwds)(Proc) — Make sure Proc subclasses are singletons</>__instancecheck__(cls,instance)— Override for isinstance(instance, cls).</>__repr__(cls)(str) — Representation for the Proc subclasses</>__subclasscheck__(cls,subclass)— Override for issubclass(subclass, cls).</>register(cls,subclass)— Register a virtual subclass of an ABC.</>
register(cls, subclass)Register a virtual subclass of an ABC.
Returns the subclass, to allow usage as a class decorator.
__instancecheck__(cls, instance)Override for isinstance(instance, cls).
__subclasscheck__(cls, subclass)Override for issubclass(subclass, cls).
__repr__(cls) → strRepresentation for the Proc subclasses
__call__(cls, *args, **kwds)Make sure Proc subclasses are singletons
*args(Any) — and**kwds(Any) — Arguments for the constructor
The Proc instance
from_proc(proc, name=None, desc=None, envs=None, envs_depth=None, cache=None, export=None, error_strategy=None, num_retries=None, forks=None, input_data=None, order=None, plugin_opts=None, requires=None, scheduler=None, scheduler_opts=None, submission_batch=None)
Create a subclass of Proc using another Proc subclass or Proc itself
proc(Type) — The Proc subclassname(str, optional) — The new name of the processdesc(str, optional) — The new description of the processenvs(Mapping, optional) — The arguments of the process, will overwrite parent oneThe items that are specified will be inheritedenvs_depth(int, optional) — How deep to update the envs when subclassed.cache(bool, optional) — Whether we should check the cache for the jobsexport(bool, optional) — When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processeserror_strategy(str, optional) — How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
num_retries(int, optional) — How many times to retry to jobs once error occursforks(int, optional) — New forks for the new processinput_data(Any, optional) — The input data for the process. Only when this processis a start processorder(int, optional) — The order to execute the new processplugin_opts(Mapping, optional) — The new plugin options, unspecified items will beinherited.requires(Sequence, optional) — The required processes for the new processscheduler(str, optional) — The new shedular to run the new processscheduler_opts(Mapping, optional) — The new scheduler options, unspecified items willbe inherited.submission_batch(int, optional) — How many jobs to be submited simultaneously.
The new process class
__init_subclass__()
Do the requirements inferring since we need them to build up theprocess relationship
run()
Init all other properties and jobs
gc()
GC process for the process to save memory after it's done
log(level, msg, *args, logger=<LoggerAdapter pipen.core (WARNING)>)
Log message for the process
level(int | str) — The log level of the recordmsg(str) — The message to log*args— The arguments to format the messagelogger(LoggerAdapter, optional) — The logging logger
biopipen.ns.tcr.ImmunarchFilter(*args, **kwds) → Proc
Immunarch - Filter data
See https://immunarch.com/articles/web_only/repFilter_v3.html
cache— Should we detect whether the jobs are cached?desc— The description of the process. Will use the summary fromthe docstring by default.dirsig— When checking the signature for caching, whether should we walkthrough the content of the directory? This is sometimes time-consuming if the directory is big.envs— The arguments that are job-independent, useful for common optionsacross jobs.envs_depth— How deep to update the envs when subclassed.error_strategy— How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
export— When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processesforks— How many jobs to run simultaneously?input— The keys for the input channelinput_data— The input data (will be computed for dependent processes)lang— The language for the script to run. Should be the path to theinterpreter iflangis not in$PATH.name— The name of the process. Will use the class name by default.nexts— Computed fromrequiresto build the process relationshipsnum_retries— How many times to retry to jobs once error occursorder— The execution order for this process. The bigger the numberis, the later the process will be executed. Default: 0. Note that the dependent processes will always be executed first. This doesn't work for start processes either, whose orders are determined byPipen.set_starts()output— The output keys for the output channel(the data will be computed)output_data— The output data (to pass to the next processes)plugin_opts— Options for process-level pluginsrequires— The dependency processesscheduler— The scheduler to run the jobsscheduler_opts— The options for the schedulerscript— The script template for the processsubmission_batch— How many jobs to be submited simultaneously.The program entrance for some schedulers may take too much resources when submitting a job or checking the job status. So we may use a smaller number here to limit the simultaneous submissions.template— Define the template engine to use.This could be either a template engine or a dict with keyengineindicating the template engine and the rest the arguments passed to the constructor of thepipen.template.Templateobject. The template engine could be either the name of the engine, currently jinja2 and liquidpy are supported, or a subclass ofpipen.template.Template. You can subclasspipen.template.Templateto use your own template engine.
filterfile— A config file in TOML.A dict of configurations with keys as the names of the group and values dicts with following keys. Seeenvs.filtersimmdata— The data loaded byimmunarch::repLoad()
groupfile— Also a group file with rownames as cells and column names aseach of the keys inin.filterfileorenvs.filters. The values will be subkeys of the dicts inin.filterfileorenvs.filters.outfile— The filteredimmdata
filters— The filters to filter the dataYou can have multiple cases (groups), the names will be the keys of this dict, values are also dicts with keys the methods supported byimmunarch::repFilter(). There is one more methodby.countsupported to filter the count matrix. Forby.meta,by.repertoire,by.rep,by.clonotypeorby.colthe values will be passed to.queryofrepFilter(). You can also use the helper functions provided byimmunarch, includingmorethan,lessthan,include,excludeandinterval. If these functions are not used,include(value)will be used by default. Forby.count, the value offilterwill be passed todplyr::filter()to filter the count matrix. You can also specifyORDERto define the filtration order, which defaults to 0, higherORDERgets later executed. Each subkey/subgroup must be exclusive For example:
{ "name": "BM_Post_Clones", "filters" { "Top_20": { "SAVE": True, # Save the filtered data to immdata "by.meta": {"Source": "BM", "Status": "Post"}, "by.count": { "ORDER": 1, "filter": "TOTAL %%in%% TOTAL[1:20]" } }, "Rest": { "by.meta": {"Source": "BM", "Status": "Post"}, "by.count": { "ORDER": 1, "filter": "!TOTAL %%in%% TOTAL[1:20]" } } }
metacols— The extra columns to be exported to the group file.prefix— The prefix will be added to the cells in the output filePlaceholders like{Sample}_can be used to from the meta data
__init_subclass__()— Do the requirements inferring since we need them to build up theprocess relationship </>from_proc(proc,name,desc,envs,envs_depth,cache,export,error_strategy,num_retries,forks,input_data,order,plugin_opts,requires,scheduler,scheduler_opts,submission_batch)(Type) — Create a subclass of Proc using another Proc subclass or Proc itself</>gc()— GC process for the process to save memory after it's done</>log(level,msg,*args,logger)— Log message for the process</>run()— Init all other properties and jobs</>
pipen.proc.ProcMeta(name, bases, namespace, **kwargs)
Meta class for Proc
__call__(cls,*args,**kwds)(Proc) — Make sure Proc subclasses are singletons</>__instancecheck__(cls,instance)— Override for isinstance(instance, cls).</>__repr__(cls)(str) — Representation for the Proc subclasses</>__subclasscheck__(cls,subclass)— Override for issubclass(subclass, cls).</>register(cls,subclass)— Register a virtual subclass of an ABC.</>
register(cls, subclass)Register a virtual subclass of an ABC.
Returns the subclass, to allow usage as a class decorator.
__instancecheck__(cls, instance)Override for isinstance(instance, cls).
__subclasscheck__(cls, subclass)Override for issubclass(subclass, cls).
__repr__(cls) → strRepresentation for the Proc subclasses
__call__(cls, *args, **kwds)Make sure Proc subclasses are singletons
*args(Any) — and**kwds(Any) — Arguments for the constructor
The Proc instance
from_proc(proc, name=None, desc=None, envs=None, envs_depth=None, cache=None, export=None, error_strategy=None, num_retries=None, forks=None, input_data=None, order=None, plugin_opts=None, requires=None, scheduler=None, scheduler_opts=None, submission_batch=None)
Create a subclass of Proc using another Proc subclass or Proc itself
proc(Type) — The Proc subclassname(str, optional) — The new name of the processdesc(str, optional) — The new description of the processenvs(Mapping, optional) — The arguments of the process, will overwrite parent oneThe items that are specified will be inheritedenvs_depth(int, optional) — How deep to update the envs when subclassed.cache(bool, optional) — Whether we should check the cache for the jobsexport(bool, optional) — When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processeserror_strategy(str, optional) — How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
num_retries(int, optional) — How many times to retry to jobs once error occursforks(int, optional) — New forks for the new processinput_data(Any, optional) — The input data for the process. Only when this processis a start processorder(int, optional) — The order to execute the new processplugin_opts(Mapping, optional) — The new plugin options, unspecified items will beinherited.requires(Sequence, optional) — The required processes for the new processscheduler(str, optional) — The new shedular to run the new processscheduler_opts(Mapping, optional) — The new scheduler options, unspecified items willbe inherited.submission_batch(int, optional) — How many jobs to be submited simultaneously.
The new process class
__init_subclass__()
Do the requirements inferring since we need them to build up theprocess relationship
run()
Init all other properties and jobs
gc()
GC process for the process to save memory after it's done
log(level, msg, *args, logger=<LoggerAdapter pipen.core (WARNING)>)
Log message for the process
level(int | str) — The log level of the recordmsg(str) — The message to log*args— The arguments to format the messagelogger(LoggerAdapter, optional) — The logging logger
biopipen.ns.tcr.Immunarch(*args, **kwds) → Proc
Exploration of Single-cell and Bulk T-cell/Antibody Immune Repertoires
See https://immunarch.com/articles/web_only/v3_basic_analysis.html
After ImmunarchLoading loads the raw data into an immunarch object,
this process wraps the functions from immunarch to do the following:
- Basic statistics, provided by
immunarch::repExplore, such as number of clones or distributions of lengths and counts. - The clonality of repertoires, provided by
immunarch::repClonality - The repertoire overlap, provided by
immunarch::repOverlap - The repertoire overlap, including different clustering procedures and PCA, provided by
immunarch::repOverlapAnalysis - The distributions of V or J genes, provided by
immunarch::geneUsage - The diversity of repertoires, provided by
immunarch::repDiversity - The dynamics of repertoires across time points/samples, provided by
immunarch::trackClonotypes - The spectratype of clonotypes, provided by
immunarch::spectratype - The distributions of kmers and sequence profiles, provided by
immunarch::getKmers - The V-J junction circos plots, implemented within the script of this process.
Environment Variable Design:
With different sets of arguments, a single function of the above can perform different tasks.
For example, repExplore can be used to get the statistics of the size of the repertoire,
the statistics of the length of the CDR3 region, or the statistics of the number of
the clonotypes. Other than that, you can also have different ways to visualize the results,
by passing different arguments to the immunarch::vis function.
For example, you can pass .by to vis to visualize the results of repExplore by different groups.
Before we explain each environment variable in details in the next section, we will give some examples here to show how the environment variables are organized in order for a single function to perform different tasks.
```toml
# Repertoire overlapping
[Immunarch.envs.overlaps]
# The method to calculate the overlap, passed to `repOverlap`
method = "public"
```
What if we want to calculate the overlap by different methods at the same time? We can use the following configuration:
```toml
[Immunarch.envs.overlaps.cases]
Public = { method = "public" }
Jaccard = { method = "jaccard" }
```
Then, the `repOverlap` function will be called twice, once with `method = "public"` and once with `method = "jaccard"`. We can also use different arguments to visualize the results. These arguments will be passed to the `vis` function:
```toml
[Immunarch.envs.overlaps.cases.Public]
method = "public"
vis_args = { "-plot": "heatmap2" }
[Immunarch.envs.overlaps.cases.Jaccard]
method = "jaccard"
vis_args = { "-plot": "heatmap2" }
```
`-plot` will be translated to `.plot` and then passed to `vis`.
If multiple cases share the same arguments, we can use the following configuration:
```toml
[Immunarch.envs.overlaps]
vis_args = { "-plot": "heatmap2" }
[Immunarch.envs.overlaps.cases]
Public = { method = "public" }
Jaccard = { method = "jaccard" }
```
For some results, there are futher analysis that can be performed. For example, for the repertoire overlap, we can perform clustering and PCA (see also <https://immunarch.com/articles/web_only/v4_overlap.html>):
```R
imm_ov1 <- repOverlap(immdata$data, .method = "public", .verbose = F)
repOverlapAnalysis(imm_ov1, "mds") %>% vis()
repOverlapAnalysis(imm_ov1, "tsne") %>% vis()
```
In such a case, we can use the following configuration:
```toml
[Immunarch.envs.overlaps]
method = "public"
[Immunarch.envs.overlaps.analyses.cases]
MDS = { "-method": "mds" }
TSNE = { "-method": "tsne" }
```
Then, the `repOverlapAnalysis` function will be called twice on the result generated by `repOverlap(immdata$data, .method = "public")`, once with `.method = "mds"` and once with `.method = "tsne"`. We can also use different arguments to visualize the results. These arguments will be passed to the `vis` function:
```toml
[Immunarch.envs.overlaps]
method = "public"
[Immunarch.envs.overlaps.analyses]
# See: <https://immunarch.com/reference/vis.immunr_hclust.html>
vis_args = { "-plot": "best" }
[Immunarch.envs.overlaps.analyses.cases]
MDS = { "-method": "mds" }
TSNE = { "-method": "tsne" }
```
Generally, you don't need to specify `cases` if you only have one case. A default case will be created for you. For multiple cases, the arguments at the same level as `cases` will be inherited by all cases.
cache— Should we detect whether the jobs are cached?desc— The description of the process. Will use the summary fromthe docstring by default.dirsig— When checking the signature for caching, whether should we walkthrough the content of the directory? This is sometimes time-consuming if the directory is big.envs— The arguments that are job-independent, useful for common optionsacross jobs.envs_depth— How deep to update the envs when subclassed.error_strategy— How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
export— When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processesforks— How many jobs to run simultaneously?input— The keys for the input channelinput_data— The input data (will be computed for dependent processes)lang— The language for the script to run. Should be the path to theinterpreter iflangis not in$PATH.name— The name of the process. Will use the class name by default.nexts— Computed fromrequiresto build the process relationshipsnum_retries— How many times to retry to jobs once error occursorder— The execution order for this process. The bigger the numberis, the later the process will be executed. Default: 0. Note that the dependent processes will always be executed first. This doesn't work for start processes either, whose orders are determined byPipen.set_starts()output— The output keys for the output channel(the data will be computed)output_data— The output data (to pass to the next processes)plugin_opts— Options for process-level pluginsrequires— The dependency processesscheduler— The scheduler to run the jobsscheduler_opts— The options for the schedulerscript— The script template for the processsubmission_batch— How many jobs to be submited simultaneously.The program entrance for some schedulers may take too much resources when submitting a job or checking the job status. So we may use a smaller number here to limit the simultaneous submissions.template— Define the template engine to use.This could be either a template engine or a dict with keyengineindicating the template engine and the rest the arguments passed to the constructor of thepipen.template.Templateobject. The template engine could be either the name of the engine, currently jinja2 and liquidpy are supported, or a subclass ofpipen.template.Template. You can subclasspipen.template.Templateto use your own template engine.
[Immunarch.envs.kmers]k = 5
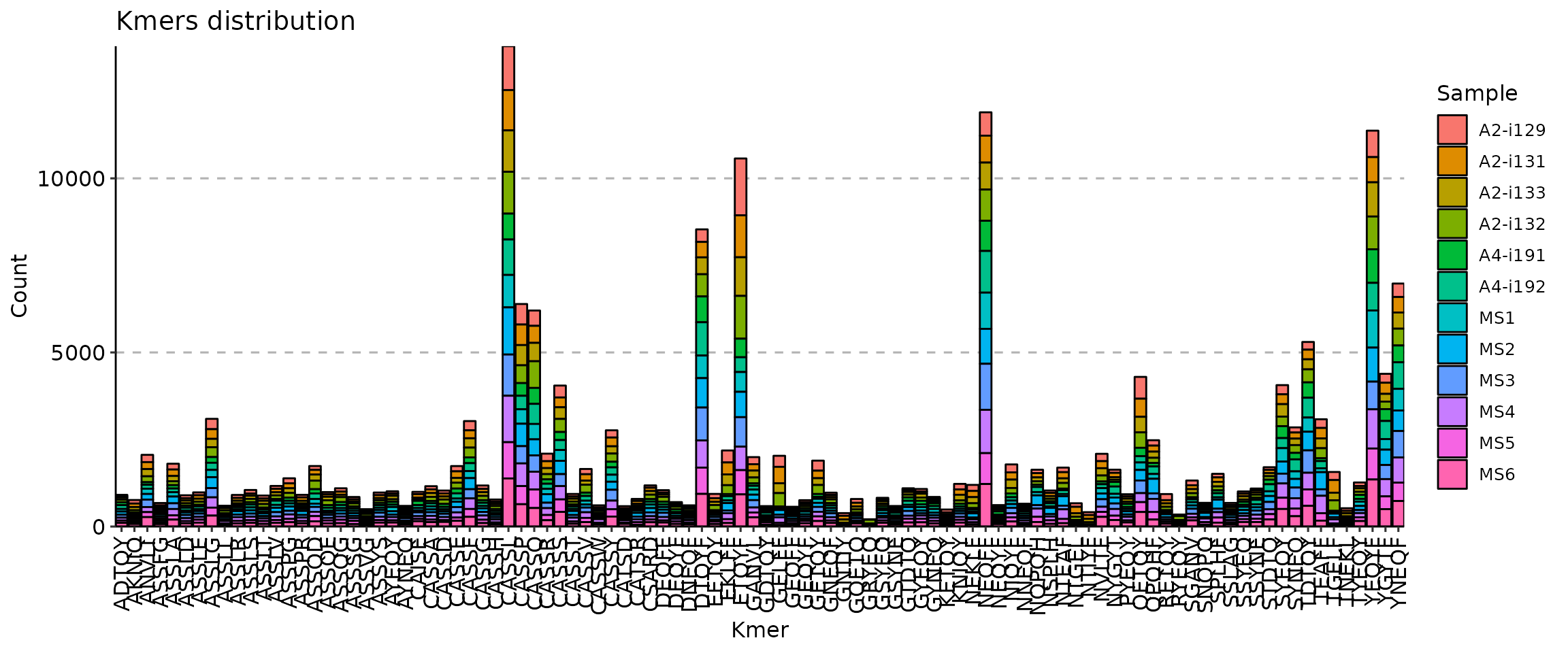
[Immunarch.envs.kmers]
# Shared by cases
k = 5
[Immunarch.envs.kmers.cases]
Head5 = { head = 5, -position = "stack" }
Head10 = { head = 10, -position = "fill" }
Head30 = { head = 30, -position = "dodge" }
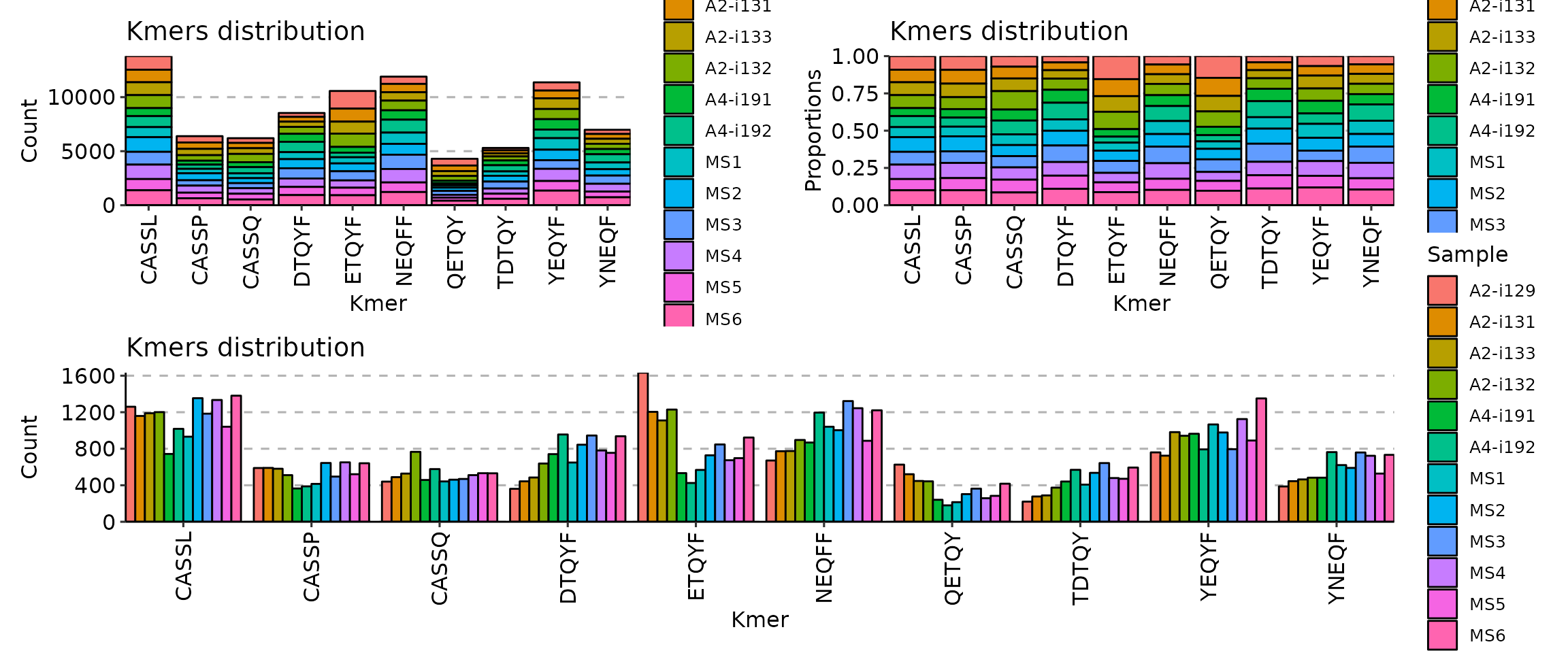
With motif profiling:
[Immunarch.envs.kmers]
k = 5
[Immnuarch.envs.kmers.profiles.cases]
TextPlot = { method = "self", vis_args = { "-plot": "text" } }
SeqPlot = { method = "self", vis_args = { "-plot": "seq" } }
immdata— The data loaded byimmunarch::repLoad()metafile— A cell-level metafile, where the first column must be the cell barcodesthat match the cell barcodes inimmdata. The other columns can be any metadata that you want to use for the analysis. The loaded metadata will be left-joined to the converted cell-level data fromimmdata. This can also be a Seurat object RDS file. If so, thesobj@meta.datawill be used as the metadata.
outdir— The output directory
counts(ns) — Explore clonotype counts.- - by: Groupings when visualize clonotype counts, passed to the
.byargument ofvis(imm_count, .by = <values>).
Multiple columns should be separated by,. - - devpars (ns): The parameters for the plotting device.
- width (type=int): The width of the plot.
- height (type=int): The height of the plot.
- res (type=int): The resolution of the plot. - - subset: Subset the data before calculating the clonotype volumes.
The whole data will be expanded to cell level, and then subsetted.
Clone sizes will be re-calculated based on the subsetted data. - - cases (type=json;order=9): If you have multiple cases, you can use this argument to specify them.
The keys will be the names of the cases.
The values will be passed to the corresponding arguments above.
If any of these arguments are not specified, the values inenvs.countswill be used.
If NO cases are specified, the default case will be added, with the nameDEFAULTand the
values ofenvs.counts.by,envs.counts.devpars.
- - by: Groupings when visualize clonotype counts, passed to the
divs(ns) — Parameters to control the diversity analysis.- - method (choice): The method to calculate diversity.
- chao1: a nonparameteric asymptotic estimator of species richness.
(number of species in a population).
- hill: Hill numbers are a mathematically unified family of diversity indices.
(differing only by an exponent q).
- div: true diversity, or the effective number of types.
It refers to the number of equally abundant types needed for the average proportional abundance of the types to equal
that observed in the dataset of interest where all types may not be equally abundant.
- gini.simp: The Gini-Simpson index.
It is the probability of interspecific encounter, i.e., probability that two entities represent different types.
- inv.simp: Inverse Simpson index.
It is the effective number of types that is obtained when the weighted arithmetic mean is used to quantify
average proportional abundance of types in the dataset of interest.
- gini: The Gini coefficient.
It measures the inequality among values of a frequency distribution (for example levels of income).
A Gini coefficient of zero expresses perfect equality, where all values are the same (for example, where everyone has the same income).
A Gini coefficient of one (or 100 percents) expresses maximal inequality among values (for example where only one person has all the income).
- d50: The D50 index.
It is the number of types that are needed to cover 50%% of the total abundance.
- raref: Species richness from the results of sampling through extrapolation. - - by: The variables (column names) to group samples.
Multiple columns should be separated by,. - - plot_type (choice): The type of the plot, works when
byis specified.
Not working forraref.
- box: Boxplot
- bar: Barplot with error bars - - subset: Subset the data before calculating the clonotype volumes.
The whole data will be expanded to cell level, and then subsetted.
Clone sizes will be re-calculated based on the subsetted data. - - args (type=json): Other arguments for
repDiversity().
Do not include the preceding.and use-instead of.in the argument names.
For example,do-normwill be compiled to.do.norm.
See all arguments at
https://immunarch.com/reference/repDiversity.html. - - order (list): The order of the values in
byon the x-axis of the plots.
If not specified, the values will be used as-is. - - test (ns): Perform statistical tests between each pair of groups.
Does NOT work forraref.
- method (choice): The method to perform the test
- none: No test
- t.test: Welch's t-test
- wilcox.test: Wilcoxon rank sum test
- padjust (choice): The method to adjust p-values.
Defaults tonone.
- bonferroni: one-step correction
- holm: step-down method using Bonferroni adjustments
- hochberg: step-up method (independent)
- hommel: closed method based on Simes tests (non-negative)
- BH: Benjamini & Hochberg (non-negative)
- BY: Benjamini & Yekutieli (negative)
- fdr: Benjamini & Hochberg (non-negative)
- none: no correction. - - separate_by: A column name used to separate the samples into different plots.
- - split_by: A column name used to split the samples into different subplots.
Likeseparate_by, but the plots will be put in the same figure.
y-axis will be shared, even ifalign_yisFalseorymin/ymaxare not specified.
ncolwill be ignored. - - split_order: The order of the values in
split_byon the x-axis of the plots.
It can also be used forseparate_byto control the order of the plots.
Values can be separated by,. - - align_x (flag): Align the x-axis of multiple plots. Only works for
raref. - - align_y (flag): Align the y-axis of multiple plots.
- - ymin (type=float): The minimum value of the y-axis.
The minimum value of the y-axis for plots splitting byseparate_by.
align_yis forcedTruewhen bothyminandymaxare specified. - - ymax (type=float): The maximum value of the y-axis.
The maximum value of the y-axis for plots splitting byseparate_by.
align_yis forcedTruewhen bothyminandymaxare specified.
Works when bothyminandymaxare specified. - - log (flag): Indicate whether we should plot with log-transformed x-axis using
vis(.log = TRUE). Only works forraref. - - ncol (type=int): The number of columns of the plots.
- - devpars (ns): The parameters for the plotting device.
- width (type=int): The width of the device
- height (type=int): The height of the device
- res (type=int): The resolution of the device - - cases (type=json;order=9): If you have multiple cases, you can use this argument to specify them.
The keys will be used as the names of the cases.
The values will be passed to the corresponding arguments above.
If NO cases are specified, the default case will be added, with the name ofenvs.div.method.
The values specified inenvs.divwill be used as the defaults for the cases here.
- - method (choice): The method to calculate diversity.
gene_usages(ns) — Explore gene usages.- - top (type=int): How many top (ranked by total usage across samples) genes to show in the plots.
Use0to use all genes. - - norm (flag): If True then use proportions of genes, else use counts of genes.
- - by: Groupings to show gene usages, passed to the
.byargument ofvis(imm_gu_top, .by = <values>).
Multiple columns should be separated by,. - - vis_args (type=json): Other arguments for the plotting functions.
- - devpars (ns): The parameters for the plotting device.
- width (type=int): The width of the plot.
- height (type=int): The height of the plot.
- res (type=int): The resolution of the plot. - - subset: Subset the data before calculating the clonotype volumes.
The whole data will be expanded to cell level, and then subsetted.
Clone sizes will be re-calculated based on the subsetted data. - - analyses (ns;order=8): Perform gene usage analyses.
- method: The method to control how the data is going to be preprocessed and analysed.
One ofjs,cor,cosine,pca,mdsandtsne. Can also be combined with following methods
for the actual analyses:hclust,kmeans,dbscan, andkruskal. For example:cosine+hclust.
You can also set tononeto skip the analyses.
See https://immunarch.com/articles/web_only/v5_gene_usage.html.
- vis_args (type=json): Other arguments for the plotting functions.
- devpars (ns): The parameters for the plotting device.
- width (type=int): The width of the plot.
- height (type=int): The height of the plot.
- res (type=int): The resolution of the plot.
- cases (type=json): If you have multiple cases, you can use this argument to specify them.
The keys will be the names of the cases.
The values will be passed to the corresponding arguments above.
If any of these arguments are not specified, the values inenvs.gene_usages.analyseswill be used.
If NO cases are specified, the default case will be added, with the nameDEFAULTand the
values ofenvs.gene_usages.analyses.method,envs.gene_usages.analyses.vis_argsandenvs.gene_usages.analyses.devpars. - - cases (type=json;order=9): If you have multiple cases, you can use this argument to specify them.
The keys will be used as the names of the cases.
The values will be passed to the corresponding arguments above.
If any of these arguments are not specified, the values inenvs.gene_usageswill be used.
If NO cases are specified, the default case will be added, with the nameDEFAULTand the
values ofenvs.gene_usages.top,envs.gene_usages.norm,envs.gene_usages.by,envs.gene_usages.vis_args,envs.gene_usages.devparsandenvs.gene_usages.analyses.
- - top (type=int): How many top (ranked by total usage across samples) genes to show in the plots.
hom_clones(ns) — Explore homeo clonotypes.- - by: Groupings when visualize homeo clones, passed to the
.byargument ofvis(imm_hom, .by = <values>).
Multiple columns should be separated by,. - - marks (ns): A dict with the threshold of the half-closed intervals that mark off clonal groups.
Passed to the.clone.typesarguments ofrepClonoality().
The keys could be:
- Rare (type=float): the rare clonotypes
- Small (type=float): the small clonotypes
- Medium (type=float): the medium clonotypes
- Large (type=float): the large clonotypes
- Hyperexpanded (type=float): the hyperexpanded clonotypes - - subset: Subset the data before calculating the clonotype volumes.
The whole data will be expanded to cell level, and then subsetted.
Clone sizes will be re-calculated based on the subsetted data. - - devpars (ns): The parameters for the plotting device.
- width (type=int): The width of the plot.
- height (type=int): The height of the plot.
- res (type=int): The resolution of the plot. - - cases (type=json;order=9): If you have multiple cases, you can use this argument to specify them.
The keys will be the names of the cases.
The values will be passed to the corresponding arguments above.
If any of these arguments are not specified, the values inenvs.hom_cloneswill be used.
If NO cases are specified, the default case will be added, with the nameDEFAULTand the
values ofenvs.hom_clones.by,envs.hom_clones.marksandenvs.hom_clones.devpars.
- - by: Groupings when visualize homeo clones, passed to the
kmers(ns) — Arguments for kmer analysis.- - k (type=int): The length of kmer.
- - head (type=int): The number of top kmers to show.
- - vis_args (type=json): Other arguments for the plotting functions.
- - devpars (ns): The parameters for the plotting device.
- width (type=int): The width of the plot.
- height (type=int): The height of the plot.
- res (type=int): The resolution of the plot. - - subset: Subset the data before calculating the clonotype volumes.
The whole data will be expanded to cell level, and then subsetted.
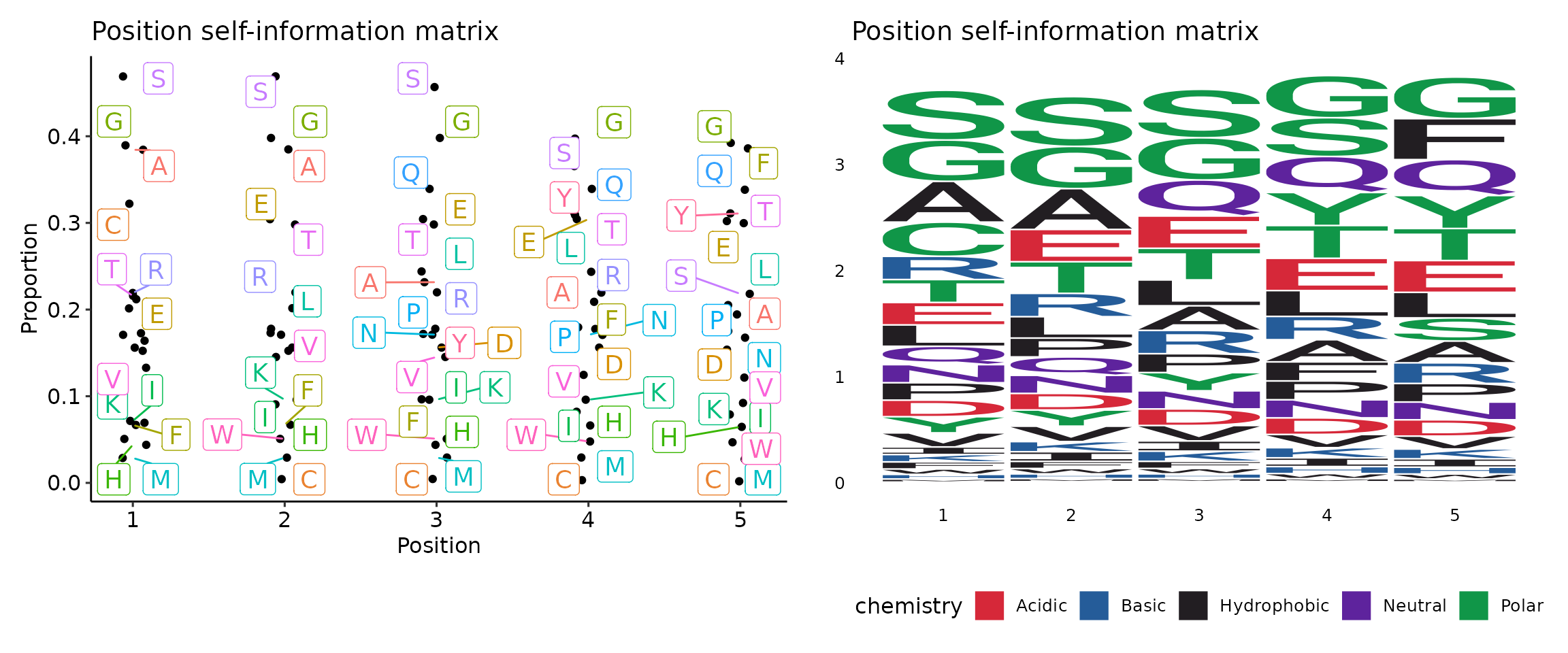
Clone sizes will be re-calculated based on the subsetted data. - - profiles (ns;order=8): Arguments for sequence profilings.
- method (choice): The method for the position matrix.
For more information see https://en.wikipedia.org/wiki/Position_weight_matrix.
- freq: position frequency matrix (PFM) - a matrix with occurences of each amino acid in each position.
- prob: position probability matrix (PPM) - a matrix with probabilities of each amino acid in each position.
- wei: position weight matrix (PWM) - a matrix with log likelihoods of PPM elements.
- self: self-information matrix (SIM) - a matrix with self-information of elements in PWM.
- vis_args (type=json): Other arguments for the plotting functions.
- devpars (ns): The parameters for the plotting device.
- width (type=int): The width of the plot.
- height (type=int): The height of the plot.
- res (type=int): The resolution of the plot.
- cases (type=json): If you have multiple cases, you can use this argument to specify them.
The keys will be the names of the cases.
The values will be passed to the corresponding arguments above.
If any of these arguments are not specified, the values inenvs.kmers.profileswill be used.
If NO cases are specified, the default case will be added, with the nameDEFAULTand the
values ofenvs.kmers.profiles.method,envs.kmers.profiles.vis_argsandenvs.kmers.profiles.devpars. - - cases (type=json;order=9): If you have multiple cases, you can use this argument to specify them.
The keys will be used as the names of the cases.
The values will be passed to the corresponding arguments above.
If any of these arguments are not specified, the default case will be added, with the nameDEFAULTand the
values ofenvs.kmers.k,envs.kmers.head,envs.kmers.vis_argsandenvs.kmers.devpars.
lens(ns) — Explore clonotype CDR3 lengths.- - by: Groupings when visualize clonotype lengths, passed to the
.byargument ofvis(imm_len, .by = <values>).
Multiple columns should be separated by,. - - devpars (ns): The parameters for the plotting device.
- width (type=int): The width of the plot.
- height (type=int): The height of the plot.
- res (type=int): The resolution of the plot. - - subset: Subset the data before calculating the clonotype volumes.
The whole data will be expanded to cell level, and then subsetted.
Clone sizes will be re-calculated based on the subsetted data. - - cases (type=json;order=9): If you have multiple cases, you can use this argument to specify them.
The keys will be the names of the cases.
The values will be passed to the corresponding arguments above.
If any of these arguments are not specified, the values inenvs.lenswill be used.
If NO cases are specified, the default case will be added, with the nameDEFAULTand the
values ofenvs.lens.by,envs.lens.devpars.
- - by: Groupings when visualize clonotype lengths, passed to the
mutaters(type=json;order=-9) — The mutaters passed todplyr::mutate()on expanded cell-level datato add new columns. The keys will be the names of the columns, and the values will be the expressions. The new names can be used involumes,lens,counts,top_clones,rare_clones,hom_clones,gene_usages,divs, etc.overlaps(ns) — Explore clonotype overlaps.- - method (choice): The method to calculate overlaps.
- public: number of public clonotypes between two samples.
- overlap: a normalised measure of overlap similarity.
It is defined as the size of the intersection divided by the smaller of the size of the two sets.
- jaccard: conceptually a percentage of how many objects two sets have in common out of how many objects they have total.
- tversky: an asymmetric similarity measure on sets that compares a variant to a prototype.
- cosine: a measure of similarity between two non-zero vectors of an inner product space that measures the cosine of the angle between them.
- morisita: how many times it is more likely to randomly select two sampled points from the same quadrat (the dataset is
covered by a regular grid of changing size) then it would be in the case of a random distribution generated from
a Poisson process. Duplicate objects are merged with their counts are summed up.
- inc+public: incremental overlaps of the N most abundant clonotypes with incrementally growing N using the public method.
- inc+morisita: incremental overlaps of the N most abundant clonotypes with incrementally growing N using the morisita method. - - subset: Subset the data before calculating the clonotype volumes.
The whole data will be expanded to cell level, and then subsetted.
Clone sizes will be re-calculated based on the subsetted data. - - vis_args (type=json): Other arguments for the plotting functions
vis(imm_ov, ...). - - devpars (ns): The parameters for the plotting device.
- width (type=int): The width of the plot.
- height (type=int): The height of the plot.
- res (type=int): The resolution of the plot. - - analyses (ns;order=8): Perform overlap analyses.
- method: Plot the samples with these dimension reduction methods.
The methods could behclust,tsne,mdsor combination of them, such asmds+hclust.
You can also set tononeto skip the analyses.
They could also be combined, for example,mds+hclust.
See https://immunarch.com/reference/repOverlapAnalysis.html.
- vis_args (type=json): Other arguments for the plotting functions.
- devpars (ns): The parameters for the plotting device.
- width (type=int): The width of the plot.
- height (type=int): The height of the plot.
- res (type=int): The resolution of the plot.
- cases (type=json): If you have multiple cases, you can use this argument to specify them.
The keys will be the names of the cases.
The values will be passed to the corresponding arguments above.
If any of these arguments are not specified, the values inenvs.overlaps.analyseswill be used.
If NO cases are specified, the default case will be added, with the nameDEFAULTand the
values ofenvs.overlaps.analyses.method,envs.overlaps.analyses.vis_argsandenvs.overlaps.analyses.devpars. - - cases (type=json;order=9): If you have multiple cases, you can use this argument to specify them.
The keys will be the names of the cases.
The values will be passed to the corresponding arguments above.
If any of these arguments are not specified, the values inenvs.overlapswill be used.
If NO cases are specified, the default case will be added, with the key the default method and the
values ofenvs.overlaps.method,envs.overlaps.vis_args,envs.overlaps.devparsandenvs.overlaps.analyses.
- - method (choice): The method to calculate overlaps.
prefix— The prefix to the barcodes. You can use placeholder like{Sample}_The prefixed barcodes will be used to match the barcodes inin.metafile. Not used ifin.metafileis not specified. IfNone(default),immdata$prefixwill be used.rare_clones(ns) — Explore rare clonotypes.- - by: Groupings when visualize rare clones, passed to the
.byargument ofvis(imm_rare, .by = <values>).
Multiple columns should be separated by,. - - marks (list;itype=int): A numerical vector with ranges of abundance for the rare clonotypes in the dataset.
Passed to the.boundargument ofrepClonoality(). - - devpars (ns): The parameters for the plotting device.
- width (type=int): The width of the plot.
- height (type=int): The height of the plot.
- res (type=int): The resolution of the plot. - - subset: Subset the data before calculating the clonotype volumes.
The whole data will be expanded to cell level, and then subsetted.
Clone sizes will be re-calculated based on the subsetted data. - - cases (type=json;order=9): If you have multiple cases, you can use this argument to specify them.
The keys will be the names of the cases.
The values will be passed to the corresponding arguments above.
If any of these arguments are not specified, the values inenvs.rare_cloneswill be used.
If NO cases are specified, the default case will be added, with the nameDEFAULTand the
values ofenvs.rare_clones.by,envs.rare_clones.marksandenvs.rare_clones.devpars.
- - by: Groupings when visualize rare clones, passed to the
spects(ns) — Spectratyping analysis.- - quant: Select the column with clonal counts to evaluate.
Set toidto count every clonotype once.
Set tocountto take into the account number of clones per clonotype.
Multiple columns should be separated by,. - - col: A string that specifies the column(s) to be processed.
The output is one of the following strings, separated by the plus sign: "nt" for nucleotide sequences,
"aa" for amino acid sequences, "v" for V gene segments, "j" for J gene segments.
E.g., pass "aa+v" for spectratyping on CDR3 amino acid sequences paired with V gene segments,
i.e., in this case a unique clonotype is a pair of CDR3 amino acid and V gene segment.
Clonal counts of equal clonotypes will be summed up. - - subset: Subset the data before calculating the clonotype volumes.
The whole data will be expanded to cell level, and then subsetted.
Clone sizes will be re-calculated based on the subsetted data. - - devpars (ns): The parameters for the plotting device.
- width (type=int): The width of the plot.
- height (type=int): The height of the plot.
- res (type=int): The resolution of the plot. - - cases (type=json;order=9): If you have multiple cases, you can use this argument to specify them.
The keys will be the names of the cases.
The values will be passed to the corresponding arguments above.
If any of these arguments are not specified, the values inenvs.spectswill be used.
By default, aBy_Clonotypecase will be added, with the values ofquant = "id"andcol = "nt", and
aBy_Num_Clonescase will be added, with the values ofquant = "count"andcol = "aa+v".
- - quant: Select the column with clonal counts to evaluate.
top_clones(ns) — Explore top clonotypes.- - by: Groupings when visualize top clones, passed to the
.byargument ofvis(imm_top, .by = <values>).
Multiple columns should be separated by,. - - marks (list;itype=int): A numerical vector with ranges of the top clonotypes. Passed to the
.headargument ofrepClonoality(). - - devpars (ns): The parameters for the plotting device.
- width (type=int): The width of the plot.
- height (type=int): The height of the plot.
- res (type=int): The resolution of the plot. - - subset: Subset the data before calculating the clonotype volumes.
The whole data will be expanded to cell level, and then subsetted.
Clone sizes will be re-calculated based on the subsetted data. - - cases (type=json;order=9): If you have multiple cases, you can use this argument to specify them.
The keys will be the names of the cases.
The values will be passed to the corresponding arguments above.
If any of these arguments are not specified, the values inenvs.top_cloneswill be used.
If NO cases are specified, the default case will be added, with the nameDEFAULTand the
values ofenvs.top_clones.by,envs.top_clones.marksandenvs.top_clones.devpars.
- - by: Groupings when visualize top clones, passed to the
trackings(ns) — Parameters to control the clonotype tracking analysis.- - targets: Either a set of CDR3AA seq of clonotypes to track (separated by
,), or simply an integer to track the top N clonotypes. - - subject_col: The column name in meta data that contains the subjects/samples on the x-axis of the alluvial plot.
If the values in this column are not unique, the values will be merged with the values insubject_colto form the x-axis.
This defaults toSample. - - subset: Subset the data before calculating the clonotype volumes.
The whole data will be expanded to cell level, and then subsetted.
Clone sizes will be re-calculated based on the subsetted data. - - subjects (list): A list of values from
subject_colto show in the alluvial plot on the x-axis.
If not specified, all values insubject_colwill be used.
This also specifies the order of the x-axis. - - cases (type=json;order=9): If you have multiple cases, you can use this argument to specify them.
The keys will be used as the names of the cases.
The values will be passed to the corresponding arguments (target,subject_col, andsubjects).
If any of these arguments are not specified, the values inenvs.trackingswill be used.
If NO cases are specified, the default case will be added, with the nameDEFAULTand the
values ofenvs.trackings.target,envs.trackings.subject_col, andenvs.trackings.subjects.
- - targets: Either a set of CDR3AA seq of clonotypes to track (separated by
vj_junc(ns) — Arguments for VJ junction circos plots.This analysis is not included inimmunarch. It is a separate implementation usingcirclize.- - by: Groupings to show VJ usages. Typically, this is the
Samplecolumn, so that the VJ usages are shown for each sample.
But you can also use other columns, such asSubjectto show the VJ usages for each subject.
Multiple columns should be separated by,. - - by_clones (flag): If True, the VJ usages will be calculated based on the distinct clonotypes, instead of the individual cells.
- - subset: Subset the data before plotting VJ usages.
The whole data will be expanded to cell level, and then subsetted.
Clone sizes will be re-calculated based on the subsetted data, which will affect the VJ usages at cell level (by_clones=False). - - devpars (ns): The parameters for the plotting device.
- width (type=int): The width of the plot.
- height (type=int): The height of the plot.
- res (type=int): The resolution of the plot. - - cases (type=json;order=9): If you have multiple cases, you can use this argument to specify them.
The keys will be used as the names of the cases. The values will be passed to the corresponding arguments above.
If any of these arguments are not specified, the values inenvs.vj_juncwill be used.
If NO cases are specified, the default case will be added, with the nameDEFAULTand the
values ofenvs.vj_junc.by,envs.vj_junc.by_clonesenvs.vj_junc.subsetandenvs.vj_junc.devpars.
- - by: Groupings to show VJ usages. Typically, this is the
volumes(ns) — Explore clonotype volume (sizes).- - by: Groupings when visualize clonotype volumes, passed to the
.byargument ofvis(imm_vol, .by = <values>).
Multiple columns should be separated by,. - - devpars (ns): The parameters for the plotting device.
- width (type=int): The width of the plot.
- height (type=int): The height of the plot.
- res (type=int): The resolution of the plot. - - subset: Subset the data before calculating the clonotype volumes.
The whole data will be expanded to cell level, and then subsetted.
Clone sizes will be re-calculated based on the subsetted data. - - cases (type=json;order=9): If you have multiple cases, you can use this argument to specify them.
The keys will be the names of the cases.
The values will be passed to the corresponding arguments above.
If any of these arguments are not specified, the values inenvs.volumeswill be used.
If NO cases are specified, the default case will be added, with the nameDEFAULTand the
values ofenvs.volume.by,envs.volume.devpars.
- - by: Groupings when visualize clonotype volumes, passed to the
__init_subclass__()— Do the requirements inferring since we need them to build up theprocess relationship </>from_proc(proc,name,desc,envs,envs_depth,cache,export,error_strategy,num_retries,forks,input_data,order,plugin_opts,requires,scheduler,scheduler_opts,submission_batch)(Type) — Create a subclass of Proc using another Proc subclass or Proc itself</>gc()— GC process for the process to save memory after it's done</>log(level,msg,*args,logger)— Log message for the process</>run()— Init all other properties and jobs</>
pipen.proc.ProcMeta(name, bases, namespace, **kwargs)
Meta class for Proc
__call__(cls,*args,**kwds)(Proc) — Make sure Proc subclasses are singletons</>__instancecheck__(cls,instance)— Override for isinstance(instance, cls).</>__repr__(cls)(str) — Representation for the Proc subclasses</>__subclasscheck__(cls,subclass)— Override for issubclass(subclass, cls).</>register(cls,subclass)— Register a virtual subclass of an ABC.</>
register(cls, subclass)Register a virtual subclass of an ABC.
Returns the subclass, to allow usage as a class decorator.
__instancecheck__(cls, instance)Override for isinstance(instance, cls).
__subclasscheck__(cls, subclass)Override for issubclass(subclass, cls).
__repr__(cls) → strRepresentation for the Proc subclasses
__call__(cls, *args, **kwds)Make sure Proc subclasses are singletons
*args(Any) — and**kwds(Any) — Arguments for the constructor
The Proc instance
from_proc(proc, name=None, desc=None, envs=None, envs_depth=None, cache=None, export=None, error_strategy=None, num_retries=None, forks=None, input_data=None, order=None, plugin_opts=None, requires=None, scheduler=None, scheduler_opts=None, submission_batch=None)
Create a subclass of Proc using another Proc subclass or Proc itself
proc(Type) — The Proc subclassname(str, optional) — The new name of the processdesc(str, optional) — The new description of the processenvs(Mapping, optional) — The arguments of the process, will overwrite parent oneThe items that are specified will be inheritedenvs_depth(int, optional) — How deep to update the envs when subclassed.cache(bool, optional) — Whether we should check the cache for the jobsexport(bool, optional) — When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processeserror_strategy(str, optional) — How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
num_retries(int, optional) — How many times to retry to jobs once error occursforks(int, optional) — New forks for the new processinput_data(Any, optional) — The input data for the process. Only when this processis a start processorder(int, optional) — The order to execute the new processplugin_opts(Mapping, optional) — The new plugin options, unspecified items will beinherited.requires(Sequence, optional) — The required processes for the new processscheduler(str, optional) — The new shedular to run the new processscheduler_opts(Mapping, optional) — The new scheduler options, unspecified items willbe inherited.submission_batch(int, optional) — How many jobs to be submited simultaneously.
The new process class
__init_subclass__()
Do the requirements inferring since we need them to build up theprocess relationship
run()
Init all other properties and jobs
gc()
GC process for the process to save memory after it's done
log(level, msg, *args, logger=<LoggerAdapter pipen.core (WARNING)>)
Log message for the process
level(int | str) — The log level of the recordmsg(str) — The message to log*args— The arguments to format the messagelogger(LoggerAdapter, optional) — The logging logger
biopipen.ns.tcr.SampleDiversity(*args, **kwds) → Proc
Sample diversity and rarefaction analysis
This is part of Immunarch, in case we have multiple dataset to compare.
cache— Should we detect whether the jobs are cached?desc— The description of the process. Will use the summary fromthe docstring by default.dirsig— When checking the signature for caching, whether should we walkthrough the content of the directory? This is sometimes time-consuming if the directory is big.envs— The arguments that are job-independent, useful for common optionsacross jobs.envs_depth— How deep to update the envs when subclassed.error_strategy— How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
export— When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processesforks— How many jobs to run simultaneously?input— The keys for the input channelinput_data— The input data (will be computed for dependent processes)lang— The language for the script to run. Should be the path to theinterpreter iflangis not in$PATH.name— The name of the process. Will use the class name by default.nexts— Computed fromrequiresto build the process relationshipsnum_retries— How many times to retry to jobs once error occursorder— The execution order for this process. The bigger the numberis, the later the process will be executed. Default: 0. Note that the dependent processes will always be executed first. This doesn't work for start processes either, whose orders are determined byPipen.set_starts()output— The output keys for the output channel(the data will be computed)output_data— The output data (to pass to the next processes)plugin_opts— Options for process-level pluginsrequires— The dependency processesscheduler— The scheduler to run the jobsscheduler_opts— The options for the schedulerscript— The script template for the processsubmission_batch— How many jobs to be submited simultaneously.The program entrance for some schedulers may take too much resources when submitting a job or checking the job status. So we may use a smaller number here to limit the simultaneous submissions.template— Define the template engine to use.This could be either a template engine or a dict with keyengineindicating the template engine and the rest the arguments passed to the constructor of thepipen.template.Templateobject. The template engine could be either the name of the engine, currently jinja2 and liquidpy are supported, or a subclass ofpipen.template.Template. You can subclasspipen.template.Templateto use your own template engine.
immdata— The data loaded byimmunarch::repLoad()
outdir— The output directory
devpars— The parameters for the plotting deviceIt is a dict, and keys are the methods and values are dicts with width, height and res that will be passed topng()If not provided, 1000, 1000 and 100 will be used.div_methods— Methods to calculate diversitiesIt is a dict, keys are the method names, values are the groupings. Each one is a case, multiple columns for a case are separated by,For example:{"div": ["Status", "Sex", "Status,Sex"]}will run true diversity for samples grouped byStatus,Sex, and both. The diversity for each sample without grouping will also be added anyway. Supported methods:chao1,hill,div,gini.simp,inv.simp,gini, andraref. See also https://immunarch.com/articles/web_only/v6_diversity.html.
__init_subclass__()— Do the requirements inferring since we need them to build up theprocess relationship </>from_proc(proc,name,desc,envs,envs_depth,cache,export,error_strategy,num_retries,forks,input_data,order,plugin_opts,requires,scheduler,scheduler_opts,submission_batch)(Type) — Create a subclass of Proc using another Proc subclass or Proc itself</>gc()— GC process for the process to save memory after it's done</>log(level,msg,*args,logger)— Log message for the process</>run()— Init all other properties and jobs</>
pipen.proc.ProcMeta(name, bases, namespace, **kwargs)
Meta class for Proc
__call__(cls,*args,**kwds)(Proc) — Make sure Proc subclasses are singletons</>__instancecheck__(cls,instance)— Override for isinstance(instance, cls).</>__repr__(cls)(str) — Representation for the Proc subclasses</>__subclasscheck__(cls,subclass)— Override for issubclass(subclass, cls).</>register(cls,subclass)— Register a virtual subclass of an ABC.</>
register(cls, subclass)Register a virtual subclass of an ABC.
Returns the subclass, to allow usage as a class decorator.
__instancecheck__(cls, instance)Override for isinstance(instance, cls).
__subclasscheck__(cls, subclass)Override for issubclass(subclass, cls).
__repr__(cls) → strRepresentation for the Proc subclasses
__call__(cls, *args, **kwds)Make sure Proc subclasses are singletons
*args(Any) — and**kwds(Any) — Arguments for the constructor
The Proc instance
from_proc(proc, name=None, desc=None, envs=None, envs_depth=None, cache=None, export=None, error_strategy=None, num_retries=None, forks=None, input_data=None, order=None, plugin_opts=None, requires=None, scheduler=None, scheduler_opts=None, submission_batch=None)
Create a subclass of Proc using another Proc subclass or Proc itself
proc(Type) — The Proc subclassname(str, optional) — The new name of the processdesc(str, optional) — The new description of the processenvs(Mapping, optional) — The arguments of the process, will overwrite parent oneThe items that are specified will be inheritedenvs_depth(int, optional) — How deep to update the envs when subclassed.cache(bool, optional) — Whether we should check the cache for the jobsexport(bool, optional) — When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processeserror_strategy(str, optional) — How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
num_retries(int, optional) — How many times to retry to jobs once error occursforks(int, optional) — New forks for the new processinput_data(Any, optional) — The input data for the process. Only when this processis a start processorder(int, optional) — The order to execute the new processplugin_opts(Mapping, optional) — The new plugin options, unspecified items will beinherited.requires(Sequence, optional) — The required processes for the new processscheduler(str, optional) — The new shedular to run the new processscheduler_opts(Mapping, optional) — The new scheduler options, unspecified items willbe inherited.submission_batch(int, optional) — How many jobs to be submited simultaneously.
The new process class
__init_subclass__()
Do the requirements inferring since we need them to build up theprocess relationship
run()
Init all other properties and jobs
gc()
GC process for the process to save memory after it's done
log(level, msg, *args, logger=<LoggerAdapter pipen.core (WARNING)>)
Log message for the process
level(int | str) — The log level of the recordmsg(str) — The message to log*args— The arguments to format the messagelogger(LoggerAdapter, optional) — The logging logger
biopipen.ns.tcr.CloneResidency(*args, **kwds) → Proc
Identification of clone residency
This process is used to investigate the residency of clones in groups, typically two samples (e.g. tumor and normal) from the same patient. But it can be used for any two groups of clones.
There are three types of output from this process
-
Count tables of the clones in the two groups
CDR3_aa Tumor Normal CASSYGLSWGSYEQYF 306 55 CASSVTGAETQYF 295 37 CASSVPSAHYNEQFF 197 9 ... ... ... -
Residency plots showing the residency of clones in the two groups
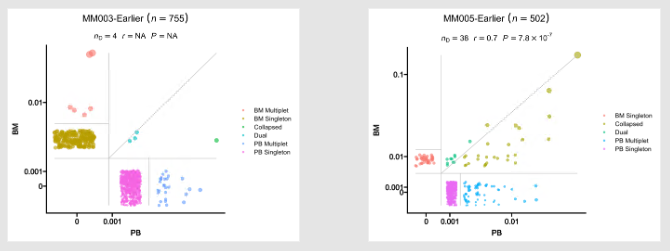
The points in the plot are jittered to avoid overplotting. The x-axis is the residency in the first group and the y-axis is the residency in the second group. The size of the points are relative to the normalized size of the clones. You may identify different types of clones in the plot based on their residency in the two groups:
- Collapsed (The clones that are collapsed in the second group)
- Dual (The clones that are present in both groups with equal size)
- Expanded (The clones that are expanded in the second group)
- First Group Multiplet (The clones only in the First Group with size > 1)
- Second Group Multiplet (The clones only in the Second Group with size > 1)
- First Group Singlet (The clones only in the First Group with size = 1)
- Second Group Singlet (The clones only in the Second Group with size = 1)
This idea is borrowed from this paper:
-
Venn diagrams showing the overlap of the clones in the two groups
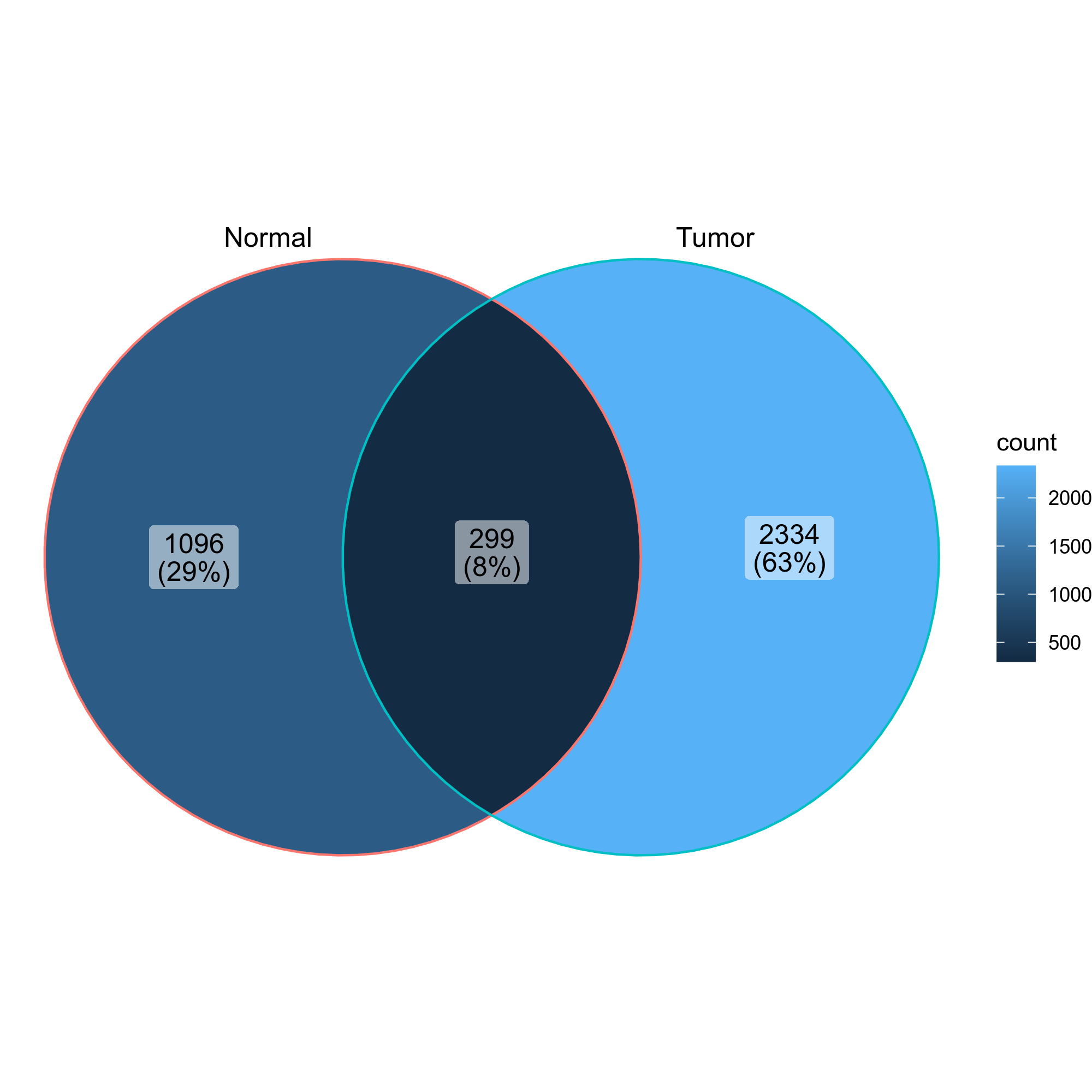 {: width="60%"}
{: width="60%"}
cache— Should we detect whether the jobs are cached?desc— The description of the process. Will use the summary fromthe docstring by default.dirsig— When checking the signature for caching, whether should we walkthrough the content of the directory? This is sometimes time-consuming if the directory is big.envs— The arguments that are job-independent, useful for common optionsacross jobs.envs_depth— How deep to update the envs when subclassed.error_strategy— How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
export— When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processesforks— How many jobs to run simultaneously?input— The keys for the input channelinput_data— The input data (will be computed for dependent processes)lang— The language for the script to run. Should be the path to theinterpreter iflangis not in$PATH.name— The name of the process. Will use the class name by default.nexts— Computed fromrequiresto build the process relationshipsnum_retries— How many times to retry to jobs once error occursorder— The execution order for this process. The bigger the numberis, the later the process will be executed. Default: 0. Note that the dependent processes will always be executed first. This doesn't work for start processes either, whose orders are determined byPipen.set_starts()output— The output keys for the output channel(the data will be computed)output_data— The output data (to pass to the next processes)plugin_opts— Options for process-level pluginsrequires— The dependency processesscheduler— The scheduler to run the jobsscheduler_opts— The options for the schedulerscript— The script template for the processsubmission_batch— How many jobs to be submited simultaneously.The program entrance for some schedulers may take too much resources when submitting a job or checking the job status. So we may use a smaller number here to limit the simultaneous submissions.template— Define the template engine to use.This could be either a template engine or a dict with keyengineindicating the template engine and the rest the arguments passed to the constructor of thepipen.template.Templateobject. The template engine could be either the name of the engine, currently jinja2 and liquidpy are supported, or a subclass ofpipen.template.Template. You can subclasspipen.template.Templateto use your own template engine.
immdata— The data loaded byimmunarch::repLoad()metafile— A cell-level metafile, where the first column must be the cell barcodesthat match the cell barcodes inimmdata. The other columns can be any metadata that you want to use for the analysis. The loaded metadata will be left-joined to the converted cell-level data fromimmdata. This can also be a Seurat object RDS file. If so, thesobj@meta.datawill be used as the metadata.
outdir— The output directory
cases(type=json) — If you have multiple cases, you can use this argumentto specify them. The keys will be used as the names of the cases. The values will be passed to the corresponding arguments. If no cases are specified, the default case will be added, with the nameDEFAULTand the values ofenvs.subject,envs.group,envs.orderandenvs.section. These values are also the defaults for the other cases.group— The key of group in metadata. This usually marks the samplesthat you want to compare. For example, Tumor vs Normal, post-treatment vs baseline It doesn't have to be 2 groups always. If there are more than 3 groups, instead of venn diagram, upset plots will be used.mutaters(type=json) — The mutaters passed todplyr::mutate()onthe cell-level data converted fromin.immdata. Ifin.metafileis provided, the mutaters will be applied to the joined data. The keys will be the names of the new columns, and the values will be the expressions. The new names can be used insubject,group,orderandsection.order(list) — The order of the values ingroup. In scatter/residency plots,XinX,Ywill be used as x-axis andYwill be used as y-axis. You can also have multiple orders. For example:["X,Y", "X,Z"]. If you only have two groups, you can setorder = ["X", "Y"], which will be the same asorder = ["X,Y"].prefix— The prefix of the cell barcodes in theSeuratobject.section— How the subjects aligned in the report. Multiple subjects withthe same value will be grouped together. Useful for cohort with large number of samples.subject(list) — The key of subject in metadata. The cloneresidency will be examined for this subject/patientsubset— The filter passed todplyr::filter()to filter the data for the cellsbefore calculating the clone residency. For example,Clones > 1to filter out singletons.upset_trans— The transformation to apply to the y axis of upset bar plots.For example,log10orsqrt. If not specified, the y axis will be plotted as is. Note that the position of the bar plots will be dodged instead of stacked when the transformation is applied. See also https://github.com/tidyverse/ggplot2/issues/3671upset_ymax— The maximum value of the y-axis in the upset bar plots.
__init_subclass__()— Do the requirements inferring since we need them to build up theprocess relationship </>from_proc(proc,name,desc,envs,envs_depth,cache,export,error_strategy,num_retries,forks,input_data,order,plugin_opts,requires,scheduler,scheduler_opts,submission_batch)(Type) — Create a subclass of Proc using another Proc subclass or Proc itself</>gc()— GC process for the process to save memory after it's done</>log(level,msg,*args,logger)— Log message for the process</>run()— Init all other properties and jobs</>
pipen.proc.ProcMeta(name, bases, namespace, **kwargs)
Meta class for Proc
__call__(cls,*args,**kwds)(Proc) — Make sure Proc subclasses are singletons</>__instancecheck__(cls,instance)— Override for isinstance(instance, cls).</>__repr__(cls)(str) — Representation for the Proc subclasses</>__subclasscheck__(cls,subclass)— Override for issubclass(subclass, cls).</>register(cls,subclass)— Register a virtual subclass of an ABC.</>
register(cls, subclass)Register a virtual subclass of an ABC.
Returns the subclass, to allow usage as a class decorator.
__instancecheck__(cls, instance)Override for isinstance(instance, cls).
__subclasscheck__(cls, subclass)Override for issubclass(subclass, cls).
__repr__(cls) → strRepresentation for the Proc subclasses
__call__(cls, *args, **kwds)Make sure Proc subclasses are singletons
*args(Any) — and**kwds(Any) — Arguments for the constructor
The Proc instance
from_proc(proc, name=None, desc=None, envs=None, envs_depth=None, cache=None, export=None, error_strategy=None, num_retries=None, forks=None, input_data=None, order=None, plugin_opts=None, requires=None, scheduler=None, scheduler_opts=None, submission_batch=None)
Create a subclass of Proc using another Proc subclass or Proc itself
proc(Type) — The Proc subclassname(str, optional) — The new name of the processdesc(str, optional) — The new description of the processenvs(Mapping, optional) — The arguments of the process, will overwrite parent oneThe items that are specified will be inheritedenvs_depth(int, optional) — How deep to update the envs when subclassed.cache(bool, optional) — Whether we should check the cache for the jobsexport(bool, optional) — When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processeserror_strategy(str, optional) — How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
num_retries(int, optional) — How many times to retry to jobs once error occursforks(int, optional) — New forks for the new processinput_data(Any, optional) — The input data for the process. Only when this processis a start processorder(int, optional) — The order to execute the new processplugin_opts(Mapping, optional) — The new plugin options, unspecified items will beinherited.requires(Sequence, optional) — The required processes for the new processscheduler(str, optional) — The new shedular to run the new processscheduler_opts(Mapping, optional) — The new scheduler options, unspecified items willbe inherited.submission_batch(int, optional) — How many jobs to be submited simultaneously.
The new process class
__init_subclass__()
Do the requirements inferring since we need them to build up theprocess relationship
run()
Init all other properties and jobs
gc()
GC process for the process to save memory after it's done
log(level, msg, *args, logger=<LoggerAdapter pipen.core (WARNING)>)
Log message for the process
level(int | str) — The log level of the recordmsg(str) — The message to log*args— The arguments to format the messagelogger(LoggerAdapter, optional) — The logging logger
biopipen.ns.tcr.Immunarch2VDJtools(*args, **kwds) → Proc
Convert immuarch format into VDJtools input formats.
This process converts the immunarch object to the
VDJtools input files,
in order to perform the VJ gene usage analysis by
VJUsage process.
This process will generally generate a tab-delimited file for each sample, with the following columns.
count: The number of reads for this clonotypefrequency: The frequency of this clonotypeCDR3nt: The nucleotide sequence of the CDR3 regionCDR3aa: The amino acid sequence of the CDR3 regionV: The V geneD: The D geneJ: The J gene
See also: https://vdjtools-doc.readthedocs.io/en/master/input.html#vdjtools-format.
This process has no environment variables.
cache— Should we detect whether the jobs are cached?desc— The description of the process. Will use the summary fromthe docstring by default.dirsig— When checking the signature for caching, whether should we walkthrough the content of the directory? This is sometimes time-consuming if the directory is big.envs— The arguments that are job-independent, useful for common optionsacross jobs.envs_depth— How deep to update the envs when subclassed.error_strategy— How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
export— When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processesforks— How many jobs to run simultaneously?input— The keys for the input channelinput_data— The input data (will be computed for dependent processes)lang— The language for the script to run. Should be the path to theinterpreter iflangis not in$PATH.name— The name of the process. Will use the class name by default.nexts— Computed fromrequiresto build the process relationshipsnum_retries— How many times to retry to jobs once error occursorder— The execution order for this process. The bigger the numberis, the later the process will be executed. Default: 0. Note that the dependent processes will always be executed first. This doesn't work for start processes either, whose orders are determined byPipen.set_starts()output— The output keys for the output channel(the data will be computed)output_data— The output data (to pass to the next processes)plugin_opts— Options for process-level pluginsrequires— The dependency processesscheduler— The scheduler to run the jobsscheduler_opts— The options for the schedulerscript— The script template for the processsubmission_batch— How many jobs to be submited simultaneously.The program entrance for some schedulers may take too much resources when submitting a job or checking the job status. So we may use a smaller number here to limit the simultaneous submissions.template— Define the template engine to use.This could be either a template engine or a dict with keyengineindicating the template engine and the rest the arguments passed to the constructor of thepipen.template.Templateobject. The template engine could be either the name of the engine, currently jinja2 and liquidpy are supported, or a subclass ofpipen.template.Template. You can subclasspipen.template.Templateto use your own template engine.
immdata— The data loaded byimmunarch::repLoad()
outdir— The output directory containing the vdjtools input for eachsample
__init_subclass__()— Do the requirements inferring since we need them to build up theprocess relationship </>from_proc(proc,name,desc,envs,envs_depth,cache,export,error_strategy,num_retries,forks,input_data,order,plugin_opts,requires,scheduler,scheduler_opts,submission_batch)(Type) — Create a subclass of Proc using another Proc subclass or Proc itself</>gc()— GC process for the process to save memory after it's done</>log(level,msg,*args,logger)— Log message for the process</>run()— Init all other properties and jobs</>
pipen.proc.ProcMeta(name, bases, namespace, **kwargs)
Meta class for Proc
__call__(cls,*args,**kwds)(Proc) — Make sure Proc subclasses are singletons</>__instancecheck__(cls,instance)— Override for isinstance(instance, cls).</>__repr__(cls)(str) — Representation for the Proc subclasses</>__subclasscheck__(cls,subclass)— Override for issubclass(subclass, cls).</>register(cls,subclass)— Register a virtual subclass of an ABC.</>
register(cls, subclass)Register a virtual subclass of an ABC.
Returns the subclass, to allow usage as a class decorator.
__instancecheck__(cls, instance)Override for isinstance(instance, cls).
__subclasscheck__(cls, subclass)Override for issubclass(subclass, cls).
__repr__(cls) → strRepresentation for the Proc subclasses
__call__(cls, *args, **kwds)Make sure Proc subclasses are singletons
*args(Any) — and**kwds(Any) — Arguments for the constructor
The Proc instance
from_proc(proc, name=None, desc=None, envs=None, envs_depth=None, cache=None, export=None, error_strategy=None, num_retries=None, forks=None, input_data=None, order=None, plugin_opts=None, requires=None, scheduler=None, scheduler_opts=None, submission_batch=None)
Create a subclass of Proc using another Proc subclass or Proc itself
proc(Type) — The Proc subclassname(str, optional) — The new name of the processdesc(str, optional) — The new description of the processenvs(Mapping, optional) — The arguments of the process, will overwrite parent oneThe items that are specified will be inheritedenvs_depth(int, optional) — How deep to update the envs when subclassed.cache(bool, optional) — Whether we should check the cache for the jobsexport(bool, optional) — When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processeserror_strategy(str, optional) — How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
num_retries(int, optional) — How many times to retry to jobs once error occursforks(int, optional) — New forks for the new processinput_data(Any, optional) — The input data for the process. Only when this processis a start processorder(int, optional) — The order to execute the new processplugin_opts(Mapping, optional) — The new plugin options, unspecified items will beinherited.requires(Sequence, optional) — The required processes for the new processscheduler(str, optional) — The new shedular to run the new processscheduler_opts(Mapping, optional) — The new scheduler options, unspecified items willbe inherited.submission_batch(int, optional) — How many jobs to be submited simultaneously.
The new process class
__init_subclass__()
Do the requirements inferring since we need them to build up theprocess relationship
run()
Init all other properties and jobs
gc()
GC process for the process to save memory after it's done
log(level, msg, *args, logger=<LoggerAdapter pipen.core (WARNING)>)
Log message for the process
level(int | str) — The log level of the recordmsg(str) — The message to log*args— The arguments to format the messagelogger(LoggerAdapter, optional) — The logging logger
biopipen.ns.tcr.ImmunarchSplitIdents(*args, **kwds) → Proc
Split the data into multiple immunarch datasets by Idents from Seurat
Note that only the cells in both the immdata and sobjfile will be
kept.
Requires immunarch >= 0.9.0 to use select_clusters()
cache— Should we detect whether the jobs are cached?desc— The description of the process. Will use the summary fromthe docstring by default.dirsig— When checking the signature for caching, whether should we walkthrough the content of the directory? This is sometimes time-consuming if the directory is big.envs— The arguments that are job-independent, useful for common optionsacross jobs.envs_depth— How deep to update the envs when subclassed.error_strategy— How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
export— When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processesforks— How many jobs to run simultaneously?input— The keys for the input channelinput_data— The input data (will be computed for dependent processes)lang— The language for the script to run. Should be the path to theinterpreter iflangis not in$PATH.name— The name of the process. Will use the class name by default.nexts— Computed fromrequiresto build the process relationshipsnum_retries— How many times to retry to jobs once error occursorder— The execution order for this process. The bigger the numberis, the later the process will be executed. Default: 0. Note that the dependent processes will always be executed first. This doesn't work for start processes either, whose orders are determined byPipen.set_starts()output— The output keys for the output channel(the data will be computed)output_data— The output data (to pass to the next processes)plugin_opts— Options for process-level pluginsrequires— The dependency processesscheduler— The scheduler to run the jobsscheduler_opts— The options for the schedulerscript— The script template for the processsubmission_batch— How many jobs to be submited simultaneously.The program entrance for some schedulers may take too much resources when submitting a job or checking the job status. So we may use a smaller number here to limit the simultaneous submissions.template— Define the template engine to use.This could be either a template engine or a dict with keyengineindicating the template engine and the rest the arguments passed to the constructor of thepipen.template.Templateobject. The template engine could be either the name of the engine, currently jinja2 and liquidpy are supported, or a subclass ofpipen.template.Template. You can subclasspipen.template.Templateto use your own template engine.
immdata— The data loaded byimmunarch::repLoad()sobjfile— The Seurat object file.You can set a different ident byIdents(sobj) <- "new_ident"to split the data by the new ident, where"new_ident"is the an existing column in meta data
outdir— The output directory containing the RDS files of the splittedimmunarch datasets
prefix— The prefix of the cell barcodes in theSeuratobject.Once could use a fixed prefix, or a placeholder with the column name in meta data. For example,"{Sample}_"will replace the placeholder with the value of the columnSamplein meta data.sample_col— The column name in meta data that contains the sample name
__init_subclass__()— Do the requirements inferring since we need them to build up theprocess relationship </>from_proc(proc,name,desc,envs,envs_depth,cache,export,error_strategy,num_retries,forks,input_data,order,plugin_opts,requires,scheduler,scheduler_opts,submission_batch)(Type) — Create a subclass of Proc using another Proc subclass or Proc itself</>gc()— GC process for the process to save memory after it's done</>log(level,msg,*args,logger)— Log message for the process</>run()— Init all other properties and jobs</>
pipen.proc.ProcMeta(name, bases, namespace, **kwargs)
Meta class for Proc
__call__(cls,*args,**kwds)(Proc) — Make sure Proc subclasses are singletons</>__instancecheck__(cls,instance)— Override for isinstance(instance, cls).</>__repr__(cls)(str) — Representation for the Proc subclasses</>__subclasscheck__(cls,subclass)— Override for issubclass(subclass, cls).</>register(cls,subclass)— Register a virtual subclass of an ABC.</>
register(cls, subclass)Register a virtual subclass of an ABC.
Returns the subclass, to allow usage as a class decorator.
__instancecheck__(cls, instance)Override for isinstance(instance, cls).
__subclasscheck__(cls, subclass)Override for issubclass(subclass, cls).
__repr__(cls) → strRepresentation for the Proc subclasses
__call__(cls, *args, **kwds)Make sure Proc subclasses are singletons
*args(Any) — and**kwds(Any) — Arguments for the constructor
The Proc instance
from_proc(proc, name=None, desc=None, envs=None, envs_depth=None, cache=None, export=None, error_strategy=None, num_retries=None, forks=None, input_data=None, order=None, plugin_opts=None, requires=None, scheduler=None, scheduler_opts=None, submission_batch=None)
Create a subclass of Proc using another Proc subclass or Proc itself
proc(Type) — The Proc subclassname(str, optional) — The new name of the processdesc(str, optional) — The new description of the processenvs(Mapping, optional) — The arguments of the process, will overwrite parent oneThe items that are specified will be inheritedenvs_depth(int, optional) — How deep to update the envs when subclassed.cache(bool, optional) — Whether we should check the cache for the jobsexport(bool, optional) — When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processeserror_strategy(str, optional) — How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
num_retries(int, optional) — How many times to retry to jobs once error occursforks(int, optional) — New forks for the new processinput_data(Any, optional) — The input data for the process. Only when this processis a start processorder(int, optional) — The order to execute the new processplugin_opts(Mapping, optional) — The new plugin options, unspecified items will beinherited.requires(Sequence, optional) — The required processes for the new processscheduler(str, optional) — The new shedular to run the new processscheduler_opts(Mapping, optional) — The new scheduler options, unspecified items willbe inherited.submission_batch(int, optional) — How many jobs to be submited simultaneously.
The new process class
__init_subclass__()
Do the requirements inferring since we need them to build up theprocess relationship
run()
Init all other properties and jobs
gc()
GC process for the process to save memory after it's done
log(level, msg, *args, logger=<LoggerAdapter pipen.core (WARNING)>)
Log message for the process
level(int | str) — The log level of the recordmsg(str) — The message to log*args— The arguments to format the messagelogger(LoggerAdapter, optional) — The logging logger
biopipen.ns.tcr.VJUsage(*args, **kwds) → Proc
Circos-style V-J usage plot displaying the frequency ofvarious V-J junctions using vdjtools.
This process performs the VJ gene usage analysis using
VDJtools.
It wraps the PlotFancyVJUsage command in VDJtools.
The output will be a V-J junction circos plot for a single sample.
Arcs correspond to different V and J segments, scaled to their frequency in sample.
Ribbons represent V-J pairings and their size is scaled to the pairing frequency
(weighted in present case).
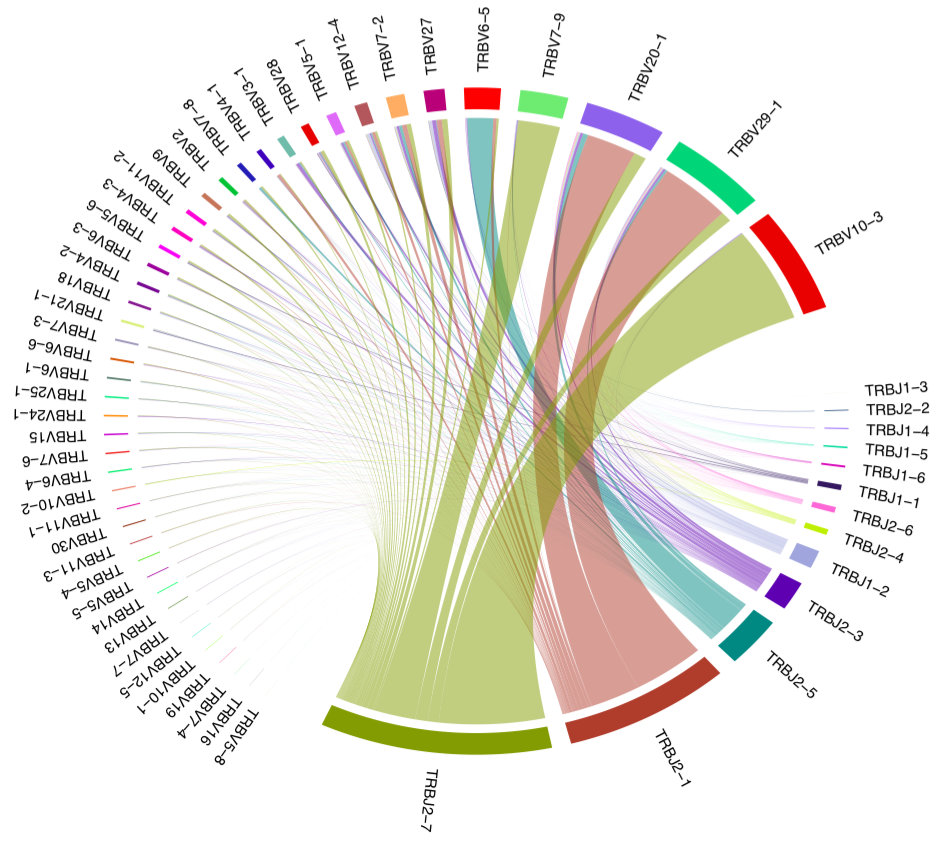 {: width="80%" }
{: width="80%" }
cache— Should we detect whether the jobs are cached?desc— The description of the process. Will use the summary fromthe docstring by default.dirsig— When checking the signature for caching, whether should we walkthrough the content of the directory? This is sometimes time-consuming if the directory is big.envs— The arguments that are job-independent, useful for common optionsacross jobs.envs_depth— How deep to update the envs when subclassed.error_strategy— How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
export— When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processesforks— How many jobs to run simultaneously?input— The keys for the input channelinput_data— The input data (will be computed for dependent processes)lang— The language for the script to run. Should be the path to theinterpreter iflangis not in$PATH.name— The name of the process. Will use the class name by default.nexts— Computed fromrequiresto build the process relationshipsnum_retries— How many times to retry to jobs once error occursorder— The execution order for this process. The bigger the numberis, the later the process will be executed. Default: 0. Note that the dependent processes will always be executed first. This doesn't work for start processes either, whose orders are determined byPipen.set_starts()output— The output keys for the output channel(the data will be computed)output_data— The output data (to pass to the next processes)plugin_opts— Options for process-level pluginsrequires— The dependency processesscheduler— The scheduler to run the jobsscheduler_opts— The options for the schedulerscript— The script template for the processsubmission_batch— How many jobs to be submited simultaneously.The program entrance for some schedulers may take too much resources when submitting a job or checking the job status. So we may use a smaller number here to limit the simultaneous submissions.template— Define the template engine to use.This could be either a template engine or a dict with keyengineindicating the template engine and the rest the arguments passed to the constructor of thepipen.template.Templateobject. The template engine could be either the name of the engine, currently jinja2 and liquidpy are supported, or a subclass ofpipen.template.Template. You can subclasspipen.template.Templateto use your own template engine.
infile— The input file, in vdjtools input format
outfile— The V-J usage plot
vdjtools— The path to theVDJtoolsexecutable.vdjtools_patch(hidden) — The patch file forVDJtools. It's delivered with the pipeline ([biopipen][3] package).- * You don't need to provide this file, unless you want to use a different patch file by yourself.
- * See the issue with
VDJtoolshere.
__init_subclass__()— Do the requirements inferring since we need them to build up theprocess relationship </>from_proc(proc,name,desc,envs,envs_depth,cache,export,error_strategy,num_retries,forks,input_data,order,plugin_opts,requires,scheduler,scheduler_opts,submission_batch)(Type) — Create a subclass of Proc using another Proc subclass or Proc itself</>gc()— GC process for the process to save memory after it's done</>log(level,msg,*args,logger)— Log message for the process</>run()— Init all other properties and jobs</>
pipen.proc.ProcMeta(name, bases, namespace, **kwargs)
Meta class for Proc
__call__(cls,*args,**kwds)(Proc) — Make sure Proc subclasses are singletons</>__instancecheck__(cls,instance)— Override for isinstance(instance, cls).</>__repr__(cls)(str) — Representation for the Proc subclasses</>__subclasscheck__(cls,subclass)— Override for issubclass(subclass, cls).</>register(cls,subclass)— Register a virtual subclass of an ABC.</>
register(cls, subclass)Register a virtual subclass of an ABC.
Returns the subclass, to allow usage as a class decorator.
__instancecheck__(cls, instance)Override for isinstance(instance, cls).
__subclasscheck__(cls, subclass)Override for issubclass(subclass, cls).
__repr__(cls) → strRepresentation for the Proc subclasses
__call__(cls, *args, **kwds)Make sure Proc subclasses are singletons
*args(Any) — and**kwds(Any) — Arguments for the constructor
The Proc instance
from_proc(proc, name=None, desc=None, envs=None, envs_depth=None, cache=None, export=None, error_strategy=None, num_retries=None, forks=None, input_data=None, order=None, plugin_opts=None, requires=None, scheduler=None, scheduler_opts=None, submission_batch=None)
Create a subclass of Proc using another Proc subclass or Proc itself
proc(Type) — The Proc subclassname(str, optional) — The new name of the processdesc(str, optional) — The new description of the processenvs(Mapping, optional) — The arguments of the process, will overwrite parent oneThe items that are specified will be inheritedenvs_depth(int, optional) — How deep to update the envs when subclassed.cache(bool, optional) — Whether we should check the cache for the jobsexport(bool, optional) — When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processeserror_strategy(str, optional) — How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
num_retries(int, optional) — How many times to retry to jobs once error occursforks(int, optional) — New forks for the new processinput_data(Any, optional) — The input data for the process. Only when this processis a start processorder(int, optional) — The order to execute the new processplugin_opts(Mapping, optional) — The new plugin options, unspecified items will beinherited.requires(Sequence, optional) — The required processes for the new processscheduler(str, optional) — The new shedular to run the new processscheduler_opts(Mapping, optional) — The new scheduler options, unspecified items willbe inherited.submission_batch(int, optional) — How many jobs to be submited simultaneously.
The new process class
__init_subclass__()
Do the requirements inferring since we need them to build up theprocess relationship
run()
Init all other properties and jobs
gc()
GC process for the process to save memory after it's done
log(level, msg, *args, logger=<LoggerAdapter pipen.core (WARNING)>)
Log message for the process
level(int | str) — The log level of the recordmsg(str) — The message to log*args— The arguments to format the messagelogger(LoggerAdapter, optional) — The logging logger
biopipen.ns.tcr.Attach2Seurat(*args, **kwds) → Proc
Attach the clonal information to a Seurat object as metadata
cache— Should we detect whether the jobs are cached?desc— The description of the process. Will use the summary fromthe docstring by default.dirsig— When checking the signature for caching, whether should we walkthrough the content of the directory? This is sometimes time-consuming if the directory is big.envs— The arguments that are job-independent, useful for common optionsacross jobs.envs_depth— How deep to update the envs when subclassed.error_strategy— How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
export— When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processesforks— How many jobs to run simultaneously?input— The keys for the input channelinput_data— The input data (will be computed for dependent processes)lang— The language for the script to run. Should be the path to theinterpreter iflangis not in$PATH.name— The name of the process. Will use the class name by default.nexts— Computed fromrequiresto build the process relationshipsnum_retries— How many times to retry to jobs once error occursorder— The execution order for this process. The bigger the numberis, the later the process will be executed. Default: 0. Note that the dependent processes will always be executed first. This doesn't work for start processes either, whose orders are determined byPipen.set_starts()output— The output keys for the output channel(the data will be computed)output_data— The output data (to pass to the next processes)plugin_opts— Options for process-level pluginsrequires— The dependency processesscheduler— The scheduler to run the jobsscheduler_opts— The options for the schedulerscript— The script template for the processsubmission_batch— How many jobs to be submited simultaneously.The program entrance for some schedulers may take too much resources when submitting a job or checking the job status. So we may use a smaller number here to limit the simultaneous submissions.template— Define the template engine to use.This could be either a template engine or a dict with keyengineindicating the template engine and the rest the arguments passed to the constructor of thepipen.template.Templateobject. The template engine could be either the name of the engine, currently jinja2 and liquidpy are supported, or a subclass ofpipen.template.Template. You can subclasspipen.template.Templateto use your own template engine.
immfile— The immunarch object in RDSsobjfile— The Seurat object file in RDS
outfile— The Seurat object with the clonal information as metadata
metacols— Which meta columns to attachprefix— The prefix to the barcodes. You can use placeholder like{Sample}_to use the meta data from the immunarch object
__init_subclass__()— Do the requirements inferring since we need them to build up theprocess relationship </>from_proc(proc,name,desc,envs,envs_depth,cache,export,error_strategy,num_retries,forks,input_data,order,plugin_opts,requires,scheduler,scheduler_opts,submission_batch)(Type) — Create a subclass of Proc using another Proc subclass or Proc itself</>gc()— GC process for the process to save memory after it's done</>log(level,msg,*args,logger)— Log message for the process</>run()— Init all other properties and jobs</>
pipen.proc.ProcMeta(name, bases, namespace, **kwargs)
Meta class for Proc
__call__(cls,*args,**kwds)(Proc) — Make sure Proc subclasses are singletons</>__instancecheck__(cls,instance)— Override for isinstance(instance, cls).</>__repr__(cls)(str) — Representation for the Proc subclasses</>__subclasscheck__(cls,subclass)— Override for issubclass(subclass, cls).</>register(cls,subclass)— Register a virtual subclass of an ABC.</>
register(cls, subclass)Register a virtual subclass of an ABC.
Returns the subclass, to allow usage as a class decorator.
__instancecheck__(cls, instance)Override for isinstance(instance, cls).
__subclasscheck__(cls, subclass)Override for issubclass(subclass, cls).
__repr__(cls) → strRepresentation for the Proc subclasses
__call__(cls, *args, **kwds)Make sure Proc subclasses are singletons
*args(Any) — and**kwds(Any) — Arguments for the constructor
The Proc instance
from_proc(proc, name=None, desc=None, envs=None, envs_depth=None, cache=None, export=None, error_strategy=None, num_retries=None, forks=None, input_data=None, order=None, plugin_opts=None, requires=None, scheduler=None, scheduler_opts=None, submission_batch=None)
Create a subclass of Proc using another Proc subclass or Proc itself
proc(Type) — The Proc subclassname(str, optional) — The new name of the processdesc(str, optional) — The new description of the processenvs(Mapping, optional) — The arguments of the process, will overwrite parent oneThe items that are specified will be inheritedenvs_depth(int, optional) — How deep to update the envs when subclassed.cache(bool, optional) — Whether we should check the cache for the jobsexport(bool, optional) — When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processeserror_strategy(str, optional) — How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
num_retries(int, optional) — How many times to retry to jobs once error occursforks(int, optional) — New forks for the new processinput_data(Any, optional) — The input data for the process. Only when this processis a start processorder(int, optional) — The order to execute the new processplugin_opts(Mapping, optional) — The new plugin options, unspecified items will beinherited.requires(Sequence, optional) — The required processes for the new processscheduler(str, optional) — The new shedular to run the new processscheduler_opts(Mapping, optional) — The new scheduler options, unspecified items willbe inherited.submission_batch(int, optional) — How many jobs to be submited simultaneously.
The new process class
__init_subclass__()
Do the requirements inferring since we need them to build up theprocess relationship
run()
Init all other properties and jobs
gc()
GC process for the process to save memory after it's done
log(level, msg, *args, logger=<LoggerAdapter pipen.core (WARNING)>)
Log message for the process
level(int | str) — The log level of the recordmsg(str) — The message to log*args— The arguments to format the messagelogger(LoggerAdapter, optional) — The logging logger
biopipen.ns.tcr.CDR3Clustering(*args, **kwds) → Proc
Cluster the TCR/BCR clones by their CDR3 sequences
This process is used to cluster TCR/BCR clones based on their CDR3 sequences.
It uses either
Zhang, Hongyi, Xiaowei Zhan, and Bo Li. "GIANA allows computationally-efficient TCR clustering and multi-disease repertoire classification by isometric transformation." Nature communications 12.1 (2021): 1-11.
Or ClusTCR
Sebastiaan Valkiers, Max Van Houcke, Kris Laukens, Pieter Meysman, ClusTCR: a Python interface for rapid clustering of large sets of CDR3 sequences with unknown antigen specificity, Bioinformatics, 2021.
Both methods are based on the Faiss Clustering Library, for efficient similarity search and clustering of dense vectors, so both methods yield similar results.
A text file will be generated with the cluster assignments for each cell, together
with the immunarch object (in R) with the cluster assignments at CDR3_Clsuter
column. This information will then be merged to a Seurat object for further
downstream analysis.
The cluster assignments are prefixed with S_ or M_ to indicate whether a
cluster has only one unique CDR3 sequence or multiple CDR3 sequences.
Note that a cluster with S_ prefix may still have multiple cells, as the same
CDR3 sequence may be shared by multiple cells.
cache— Should we detect whether the jobs are cached?desc— The description of the process. Will use the summary fromthe docstring by default.dirsig— When checking the signature for caching, whether should we walkthrough the content of the directory? This is sometimes time-consuming if the directory is big.envs— The arguments that are job-independent, useful for common optionsacross jobs.envs_depth— How deep to update the envs when subclassed.error_strategy— How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
export— When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processesforks— How many jobs to run simultaneously?input— The keys for the input channelinput_data— The input data (will be computed for dependent processes)lang— The language for the script to run. Should be the path to theinterpreter iflangis not in$PATH.name— The name of the process. Will use the class name by default.nexts— Computed fromrequiresto build the process relationshipsnum_retries— How many times to retry to jobs once error occursorder— The execution order for this process. The bigger the numberis, the later the process will be executed. Default: 0. Note that the dependent processes will always be executed first. This doesn't work for start processes either, whose orders are determined byPipen.set_starts()output— The output keys for the output channel(the data will be computed)output_data— The output data (to pass to the next processes)plugin_opts— Options for process-level pluginsrequires— The dependency processesscheduler— The scheduler to run the jobsscheduler_opts— The options for the schedulerscript— The script template for the processsubmission_batch— How many jobs to be submited simultaneously.The program entrance for some schedulers may take too much resources when submitting a job or checking the job status. So we may use a smaller number here to limit the simultaneous submissions.template— Define the template engine to use.This could be either a template engine or a dict with keyengineindicating the template engine and the rest the arguments passed to the constructor of thepipen.template.Templateobject. The template engine could be either the name of the engine, currently jinja2 and liquidpy are supported, or a subclass ofpipen.template.Template. You can subclasspipen.template.Templateto use your own template engine.
screpfile— The TCR/BCR data object loaded byscRepertoire::CombineTCR(),scRepertoire::CombineBCR()orscRepertoire::CombineExpression()
outfile— ThescRepertoireobject in qs with TCR/BCR cluster information.ColumnCDR3_Clusterwill be added to the metadata.
args(type=json) — The arguments for the clustering toolFor GIANA, they will be passed topython GIAna.pySee https://github.com/s175573/GIANA#usage. For ClusTCR, they will be passed toclustcr.Clustering(...)See https://svalkiers.github.io/clusTCR/docs/clustering/how-to-use.html#clustering.chain(choice) — The TCR/BCR chain to use for clustering.- - heavy: The heavy chain, TRB for TCR, IGH for BCR.
For TCR, TRB is the second sequence inCTaa, separated by_if
input is a Seurat object; otherwise, it is extracted from thecdr3_aa2column.
For BCR, IGH is the first sequence inCTaa, separated by_if
input is a Seurat object; otherwise, it is extracted from thecdr3_aa1column. - - light: The light chain, TRA for TCR, IGL/IGK for BCR.
For TCR, TRA is the first sequence inCTaa, separated by_if
input is a Seurat object; otherwise, it is extracted from thecdr3_aa1column.
For BCR, IGL/IGK is the second sequence inCTaa, separated by_if
input is a Seurat object; otherwise, it is extracted from thecdr3_aa2column. - - TRA: Only the TRA chain for TCR (light chain).
- - TRB: Only the TRB chain for TCR (heavy chain).
- - IGH: Only the IGH chain for BCR (heavy chain).
- - IGLK: Only the IGL/IGK chain for BCR (light chain).
- - both: Both sequences from the heavy and light chains (CTaa column).
- - heavy: The heavy chain, TRB for TCR, IGH for BCR.
python— The path of python withGIANA's dependencies installedor withclusTCRinstalled. Depending on thetoolyou choose.tool(choice) — The tool used to do the clustering, eitherGIANA or ClusTCR. For GIANA, using TRBV mutations is not supported- - GIANA: by Li lab at UT Southwestern Medical Center
- - ClusTCR: by Sebastiaan Valkiers, etc
type(choice) — The type of the data.- - TCR: T cell receptor data
- - BCR: B cell receptor data
- - auto: Automatically detect the type from the data.
Try to find TRB or IGH genes in the CTgene column to determine
whether it is TCR or BCR data.
within_sample(flag) — Whether to cluster the TCR/BCR clones within each sample.Whenin.screpfileis aSeuratobject, the samples are marked by theSamplecolumn in the metadata.
clusTCR—- if: {{ proc.envs.tool == 'ClusTCR' }}
- check: {{ proc.envs.python }} -c "import clustcr"
__init_subclass__()— Do the requirements inferring since we need them to build up theprocess relationship </>from_proc(proc,name,desc,envs,envs_depth,cache,export,error_strategy,num_retries,forks,input_data,order,plugin_opts,requires,scheduler,scheduler_opts,submission_batch)(Type) — Create a subclass of Proc using another Proc subclass or Proc itself</>gc()— GC process for the process to save memory after it's done</>log(level,msg,*args,logger)— Log message for the process</>run()— Init all other properties and jobs</>
pipen.proc.ProcMeta(name, bases, namespace, **kwargs)
Meta class for Proc
__call__(cls,*args,**kwds)(Proc) — Make sure Proc subclasses are singletons</>__instancecheck__(cls,instance)— Override for isinstance(instance, cls).</>__repr__(cls)(str) — Representation for the Proc subclasses</>__subclasscheck__(cls,subclass)— Override for issubclass(subclass, cls).</>register(cls,subclass)— Register a virtual subclass of an ABC.</>
register(cls, subclass)Register a virtual subclass of an ABC.
Returns the subclass, to allow usage as a class decorator.
__instancecheck__(cls, instance)Override for isinstance(instance, cls).
__subclasscheck__(cls, subclass)Override for issubclass(subclass, cls).
__repr__(cls) → strRepresentation for the Proc subclasses
__call__(cls, *args, **kwds)Make sure Proc subclasses are singletons
*args(Any) — and**kwds(Any) — Arguments for the constructor
The Proc instance
from_proc(proc, name=None, desc=None, envs=None, envs_depth=None, cache=None, export=None, error_strategy=None, num_retries=None, forks=None, input_data=None, order=None, plugin_opts=None, requires=None, scheduler=None, scheduler_opts=None, submission_batch=None)
Create a subclass of Proc using another Proc subclass or Proc itself
proc(Type) — The Proc subclassname(str, optional) — The new name of the processdesc(str, optional) — The new description of the processenvs(Mapping, optional) — The arguments of the process, will overwrite parent oneThe items that are specified will be inheritedenvs_depth(int, optional) — How deep to update the envs when subclassed.cache(bool, optional) — Whether we should check the cache for the jobsexport(bool, optional) — When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processeserror_strategy(str, optional) — How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
num_retries(int, optional) — How many times to retry to jobs once error occursforks(int, optional) — New forks for the new processinput_data(Any, optional) — The input data for the process. Only when this processis a start processorder(int, optional) — The order to execute the new processplugin_opts(Mapping, optional) — The new plugin options, unspecified items will beinherited.requires(Sequence, optional) — The required processes for the new processscheduler(str, optional) — The new shedular to run the new processscheduler_opts(Mapping, optional) — The new scheduler options, unspecified items willbe inherited.submission_batch(int, optional) — How many jobs to be submited simultaneously.
The new process class
__init_subclass__()
Do the requirements inferring since we need them to build up theprocess relationship
run()
Init all other properties and jobs
gc()
GC process for the process to save memory after it's done
log(level, msg, *args, logger=<LoggerAdapter pipen.core (WARNING)>)
Log message for the process
level(int | str) — The log level of the recordmsg(str) — The message to log*args— The arguments to format the messagelogger(LoggerAdapter, optional) — The logging logger
biopipen.ns.tcr.TCRClusterStats(*args, **kwds) → Proc
Statistics of TCR clusters, generated by TCRClustering.
The statistics include
- The number of cells in each cluster (cluster size)
- Sample diversity using TCR clusters instead of TCR clones
- Shared TCR clusters between samples
cache— Should we detect whether the jobs are cached?desc— The description of the process. Will use the summary fromthe docstring by default.dirsig— When checking the signature for caching, whether should we walkthrough the content of the directory? This is sometimes time-consuming if the directory is big.envs— The arguments that are job-independent, useful for common optionsacross jobs.envs_depth— How deep to update the envs when subclassed.error_strategy— How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
export— When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processesforks— How many jobs to run simultaneously?input— The keys for the input channelinput_data— The input data (will be computed for dependent processes)lang— The language for the script to run. Should be the path to theinterpreter iflangis not in$PATH.name— The name of the process. Will use the class name by default.nexts— Computed fromrequiresto build the process relationshipsnum_retries— How many times to retry to jobs once error occursorder— The execution order for this process. The bigger the numberis, the later the process will be executed. Default: 0. Note that the dependent processes will always be executed first. This doesn't work for start processes either, whose orders are determined byPipen.set_starts()output— The output keys for the output channel(the data will be computed)output_data— The output data (to pass to the next processes)plugin_opts— Options for process-level pluginsrequires— The dependency processesscheduler— The scheduler to run the jobsscheduler_opts— The options for the schedulerscript— The script template for the processsubmission_batch— How many jobs to be submited simultaneously.The program entrance for some schedulers may take too much resources when submitting a job or checking the job status. So we may use a smaller number here to limit the simultaneous submissions.template— Define the template engine to use.This could be either a template engine or a dict with keyengineindicating the template engine and the rest the arguments passed to the constructor of thepipen.template.Templateobject. The template engine could be either the name of the engine, currently jinja2 and liquidpy are supported, or a subclass ofpipen.template.Template. You can subclasspipen.template.Templateto use your own template engine.
Cluster size
[TCRClusterStats.envs.cluster_size]
by = "Sample"
 {: width="80%"}
{: width="80%"}
Shared clusters
[TCRClusterStats.envs.shared_clusters]
numbers_on_heatmap = true
heatmap_meta = ["region"]
 {: width="80%"}
{: width="80%"}
Sample diversity
[TCRClusterStats.envs.sample_diversity]
method = "gini"
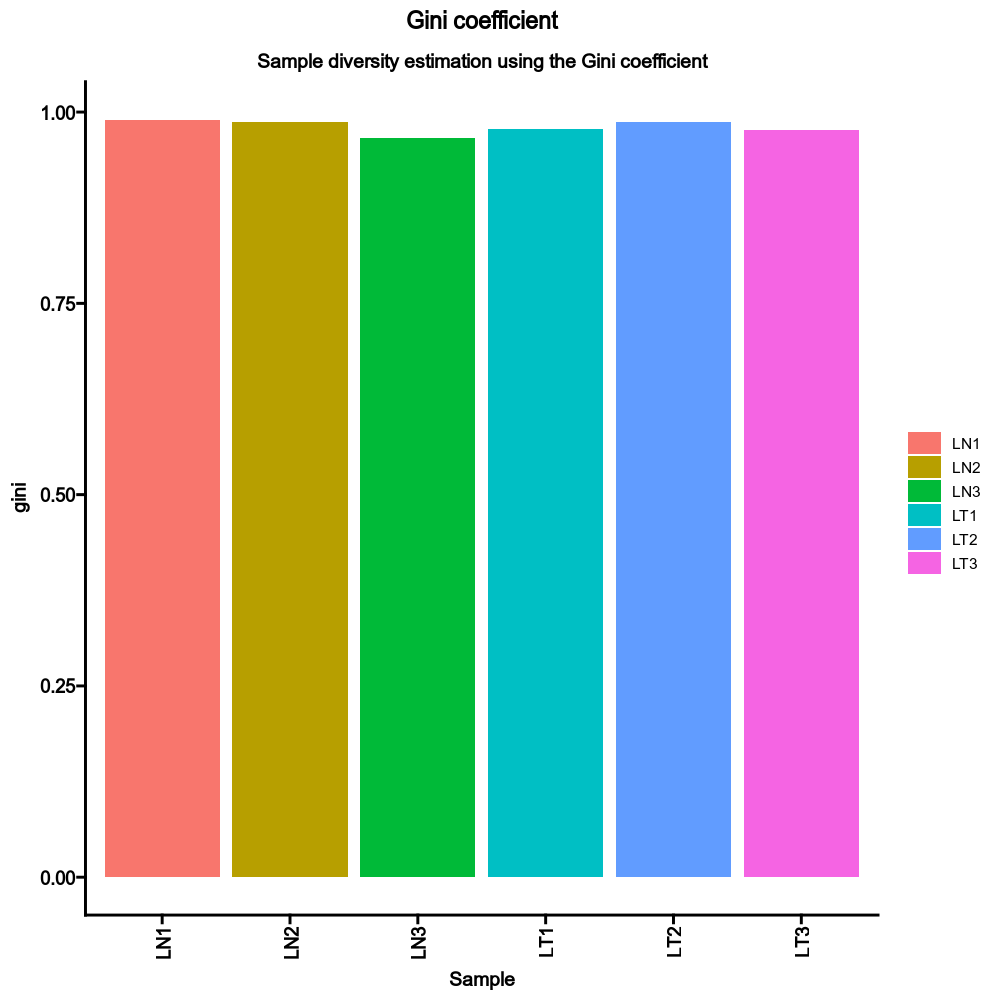 {: width="80%"}
{: width="80%"}
Compared to the sample diversity using TCR clones:
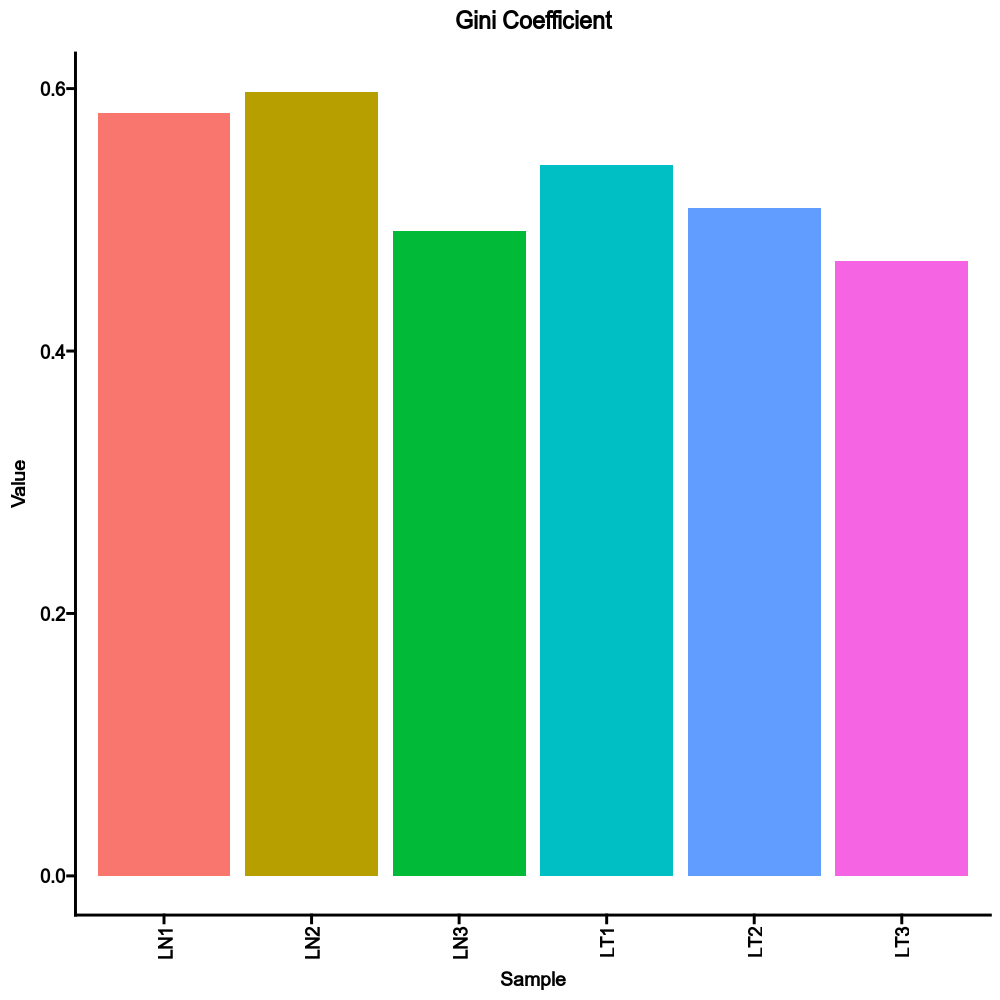 {: width="80%"}
{: width="80%"}
immfile— The immunarch object with TCR clusters attached
outdir— The output directory containing the stats and reports
cluster_size(ns) — The distribution of size of each cluster.- - by: The variables (column names) used to fill the histogram.
Only a single column is supported. - - devpars (ns): The parameters for the plotting device.
- width (type=int): The width of the device
- height (type=int): The height of the device
- res (type=int): The resolution of the device - - cases (type=json): If you have multiple cases, you can use this
argument to specify them. The keys will be the names of the
cases. The values will be passed to the corresponding arguments
above. If any of these arguments are not specified, the values
inenvs.cluster_sizewill be used. If NO cases are
specified, the default case will be added, with the name
DEFAULT.
- - by: The variables (column names) used to fill the histogram.
sample_diversity(ns) — Sample diversity using TCR clusters instead ofclones.- - by: The variables (column names) to group samples.
Multiple columns should be separated by,. - - method (choice): The method to calculate diversity.
- gini: The Gini coefficient.
It measures the inequality among values of a frequency
distribution (for example levels of income).
- gini.simp: The Gini-Simpson index.
It is the probability of interspecific encounter, i.e.,
probability that two entities represent different types.
- inv.simp: Inverse Simpson index.
It is the effective number of types that is obtained when
the weighted arithmetic mean is used to quantify average
proportional abundance of types in the dataset of interest.
- div: true diversity, or the effective number of types.
It refers to the number of equally abundant types needed
for the average proportional abundance of the types to
equal that observed in the dataset of interest where all
types may not be equally abundant. - - devpars (ns): The parameters for the plotting device.
- width (type=int): The width of the device
- height (type=int): The height of the device
- res (type=int): The resolution of the device - - cases (type=json): If you have multiple cases, you can use this
argument to specify them. The keys will be the names of the
cases. The values will be passed to the corresponding arguments
above. If any of these arguments are not specified, the values
inenvs.sample_diversitywill be used. If NO cases are
specified, the default case will be added, with the name
DEFAULT.
- - by: The variables (column names) to group samples.
shared_clusters(ns) — Stats about shared TCR clusters- - numbers_on_heatmap (flag): Whether to show the
numbers on the heatmap. - - heatmap_meta (list): The columns of metadata to show on the
heatmap. - - cluster_rows (flag): Whether to cluster the rows on the heatmap.
- - sample_order: The order of the samples on the heatmap.
Either a string separated by,or a list of sample names.
This only works for columns ifcluster_rowsisTrue. - - grouping: The groups to investigate the shared clusters.
If specified, venn diagrams will be drawn instead of heatmaps.
In such case,numbers_on_heatmapandheatmap_metawill be
ignored. - - devpars (ns): The parameters for the plotting device.
- width (type=int): The width of the device
- height (type=int): The height of the device
- res (type=int): The resolution of the device - - cases (type=json): If you have multiple cases, you can use this
argument to specify them. The keys will be the names of the
cases. The values will be passed to the corresponding arguments
above. If any of these arguments are not specified, the values
inenvs.shared_clusterswill be used. If NO cases are
specified, the default case will be added, with the name
DEFAULT.
- - numbers_on_heatmap (flag): Whether to show the
r-immunarch—- check: {{proc.lang}} -e "library(immunarch)"
__init_subclass__()— Do the requirements inferring since we need them to build up theprocess relationship </>from_proc(proc,name,desc,envs,envs_depth,cache,export,error_strategy,num_retries,forks,input_data,order,plugin_opts,requires,scheduler,scheduler_opts,submission_batch)(Type) — Create a subclass of Proc using another Proc subclass or Proc itself</>gc()— GC process for the process to save memory after it's done</>log(level,msg,*args,logger)— Log message for the process</>run()— Init all other properties and jobs</>
pipen.proc.ProcMeta(name, bases, namespace, **kwargs)
Meta class for Proc
__call__(cls,*args,**kwds)(Proc) — Make sure Proc subclasses are singletons</>__instancecheck__(cls,instance)— Override for isinstance(instance, cls).</>__repr__(cls)(str) — Representation for the Proc subclasses</>__subclasscheck__(cls,subclass)— Override for issubclass(subclass, cls).</>register(cls,subclass)— Register a virtual subclass of an ABC.</>
register(cls, subclass)Register a virtual subclass of an ABC.
Returns the subclass, to allow usage as a class decorator.
__instancecheck__(cls, instance)Override for isinstance(instance, cls).
__subclasscheck__(cls, subclass)Override for issubclass(subclass, cls).
__repr__(cls) → strRepresentation for the Proc subclasses
__call__(cls, *args, **kwds)Make sure Proc subclasses are singletons
*args(Any) — and**kwds(Any) — Arguments for the constructor
The Proc instance
from_proc(proc, name=None, desc=None, envs=None, envs_depth=None, cache=None, export=None, error_strategy=None, num_retries=None, forks=None, input_data=None, order=None, plugin_opts=None, requires=None, scheduler=None, scheduler_opts=None, submission_batch=None)
Create a subclass of Proc using another Proc subclass or Proc itself
proc(Type) — The Proc subclassname(str, optional) — The new name of the processdesc(str, optional) — The new description of the processenvs(Mapping, optional) — The arguments of the process, will overwrite parent oneThe items that are specified will be inheritedenvs_depth(int, optional) — How deep to update the envs when subclassed.cache(bool, optional) — Whether we should check the cache for the jobsexport(bool, optional) — When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processeserror_strategy(str, optional) — How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
num_retries(int, optional) — How many times to retry to jobs once error occursforks(int, optional) — New forks for the new processinput_data(Any, optional) — The input data for the process. Only when this processis a start processorder(int, optional) — The order to execute the new processplugin_opts(Mapping, optional) — The new plugin options, unspecified items will beinherited.requires(Sequence, optional) — The required processes for the new processscheduler(str, optional) — The new shedular to run the new processscheduler_opts(Mapping, optional) — The new scheduler options, unspecified items willbe inherited.submission_batch(int, optional) — How many jobs to be submited simultaneously.
The new process class
__init_subclass__()
Do the requirements inferring since we need them to build up theprocess relationship
run()
Init all other properties and jobs
gc()
GC process for the process to save memory after it's done
log(level, msg, *args, logger=<LoggerAdapter pipen.core (WARNING)>)
Log message for the process
level(int | str) — The log level of the recordmsg(str) — The message to log*args— The arguments to format the messagelogger(LoggerAdapter, optional) — The logging logger
biopipen.ns.tcr.CloneSizeQQPlot(*args, **kwds) → Proc
QQ plot of the clone sizes
QQ plots for clones sizes of pairs of samples
cache— Should we detect whether the jobs are cached?desc— The description of the process. Will use the summary fromthe docstring by default.dirsig— When checking the signature for caching, whether should we walkthrough the content of the directory? This is sometimes time-consuming if the directory is big.envs— The arguments that are job-independent, useful for common optionsacross jobs.envs_depth— How deep to update the envs when subclassed.error_strategy— How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
export— When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processesforks— How many jobs to run simultaneously?input— The keys for the input channelinput_data— The input data (will be computed for dependent processes)lang— The language for the script to run. Should be the path to theinterpreter iflangis not in$PATH.name— The name of the process. Will use the class name by default.nexts— Computed fromrequiresto build the process relationshipsnum_retries— How many times to retry to jobs once error occursorder— The execution order for this process. The bigger the numberis, the later the process will be executed. Default: 0. Note that the dependent processes will always be executed first. This doesn't work for start processes either, whose orders are determined byPipen.set_starts()output— The output keys for the output channel(the data will be computed)output_data— The output data (to pass to the next processes)plugin_opts— Options for process-level pluginsrequires— The dependency processesscheduler— The scheduler to run the jobsscheduler_opts— The options for the schedulerscript— The script template for the processsubmission_batch— How many jobs to be submited simultaneously.The program entrance for some schedulers may take too much resources when submitting a job or checking the job status. So we may use a smaller number here to limit the simultaneous submissions.template— Define the template engine to use.This could be either a template engine or a dict with keyengineindicating the template engine and the rest the arguments passed to the constructor of thepipen.template.Templateobject. The template engine could be either the name of the engine, currently jinja2 and liquidpy are supported, or a subclass ofpipen.template.Template. You can subclasspipen.template.Templateto use your own template engine.
immdata— The data loaded byimmunarch::repLoad()
outdir— The output directory
diag— Whether to draw the diagonal line in the QQ plotgroup— The key of group in metadata. This usually marks the samplesthat you want to compare. For example, Tumor vs Normal, post-treatment vs baseline It doesn't have to be 2 groups always. If there are more than 3 groups, for example, [A, B, C], the QQ plots will be generated for all the combinations of 2 groups, i.e., [A, B], [A, C], [B, C]on— The key of the metadata to use for the QQ plot. One/Both of["Clones", "Proportion"]order— The order of the values ingroup. Early-ordered group willbe used as x-axis in scatter plots If there are more than 2 groups, for example, [A, B, C], the QQ plots will be drawn for pairs: B ~ A, C ~ B.subject— The key of subject in metadata, defining the pairs.The clone residency will be examined for this subject/patient
__init_subclass__()— Do the requirements inferring since we need them to build up theprocess relationship </>from_proc(proc,name,desc,envs,envs_depth,cache,export,error_strategy,num_retries,forks,input_data,order,plugin_opts,requires,scheduler,scheduler_opts,submission_batch)(Type) — Create a subclass of Proc using another Proc subclass or Proc itself</>gc()— GC process for the process to save memory after it's done</>log(level,msg,*args,logger)— Log message for the process</>run()— Init all other properties and jobs</>
pipen.proc.ProcMeta(name, bases, namespace, **kwargs)
Meta class for Proc
__call__(cls,*args,**kwds)(Proc) — Make sure Proc subclasses are singletons</>__instancecheck__(cls,instance)— Override for isinstance(instance, cls).</>__repr__(cls)(str) — Representation for the Proc subclasses</>__subclasscheck__(cls,subclass)— Override for issubclass(subclass, cls).</>register(cls,subclass)— Register a virtual subclass of an ABC.</>
register(cls, subclass)Register a virtual subclass of an ABC.
Returns the subclass, to allow usage as a class decorator.
__instancecheck__(cls, instance)Override for isinstance(instance, cls).
__subclasscheck__(cls, subclass)Override for issubclass(subclass, cls).
__repr__(cls) → strRepresentation for the Proc subclasses
__call__(cls, *args, **kwds)Make sure Proc subclasses are singletons
*args(Any) — and**kwds(Any) — Arguments for the constructor
The Proc instance
from_proc(proc, name=None, desc=None, envs=None, envs_depth=None, cache=None, export=None, error_strategy=None, num_retries=None, forks=None, input_data=None, order=None, plugin_opts=None, requires=None, scheduler=None, scheduler_opts=None, submission_batch=None)
Create a subclass of Proc using another Proc subclass or Proc itself
proc(Type) — The Proc subclassname(str, optional) — The new name of the processdesc(str, optional) — The new description of the processenvs(Mapping, optional) — The arguments of the process, will overwrite parent oneThe items that are specified will be inheritedenvs_depth(int, optional) — How deep to update the envs when subclassed.cache(bool, optional) — Whether we should check the cache for the jobsexport(bool, optional) — When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processeserror_strategy(str, optional) — How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
num_retries(int, optional) — How many times to retry to jobs once error occursforks(int, optional) — New forks for the new processinput_data(Any, optional) — The input data for the process. Only when this processis a start processorder(int, optional) — The order to execute the new processplugin_opts(Mapping, optional) — The new plugin options, unspecified items will beinherited.requires(Sequence, optional) — The required processes for the new processscheduler(str, optional) — The new shedular to run the new processscheduler_opts(Mapping, optional) — The new scheduler options, unspecified items willbe inherited.submission_batch(int, optional) — How many jobs to be submited simultaneously.
The new process class
__init_subclass__()
Do the requirements inferring since we need them to build up theprocess relationship
run()
Init all other properties and jobs
gc()
GC process for the process to save memory after it's done
log(level, msg, *args, logger=<LoggerAdapter pipen.core (WARNING)>)
Log message for the process
level(int | str) — The log level of the recordmsg(str) — The message to log*args— The arguments to format the messagelogger(LoggerAdapter, optional) — The logging logger
biopipen.ns.tcr.CDR3AAPhyschem(*args, **kwds) → Proc
CDR3 AA physicochemical feature analysis
The idea is to perform a regression between two groups of cells (e.g. Treg vs Tconv) at different length of CDR3 AA sequences. The regression will be performed for each physicochemical feature of the AA (hydrophobicity, volume and isolectric point).
Reference:
- - Stadinski, Brian D., et al. "Hydrophobic CDR3 residues promote the development of self-reactive T cells." Nature immunology 17.8 (2016): 946-955.
- - Lagattuta, Kaitlyn A., et al. "Repertoire analyses reveal T cell antigen receptor sequence features that influence T cell fate." Nature immunology 23.3 (2022): 446-457.
- - Wimley, W. C. & White, S. H. Experimentally determined hydrophobicity scale for proteins at membrane - interfaces. Nat. Struct. Biol. 3, 842-848 (1996).
- - Handbook of chemistry & physics 72nd edition. (CRC Press, 1991).
- - Zamyatnin, A. A. Protein volume in solution. Prog. Biophys. Mol. Biol. 24, 107-123 (1972).
cache— Should we detect whether the jobs are cached?desc— The description of the process. Will use the summary fromthe docstring by default.dirsig— When checking the signature for caching, whether should we walkthrough the content of the directory? This is sometimes time-consuming if the directory is big.envs— The arguments that are job-independent, useful for common optionsacross jobs.envs_depth— How deep to update the envs when subclassed.error_strategy— How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
export— When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processesforks— How many jobs to run simultaneously?input— The keys for the input channelinput_data— The input data (will be computed for dependent processes)lang— The language for the script to run. Should be the path to theinterpreter iflangis not in$PATH.name— The name of the process. Will use the class name by default.nexts— Computed fromrequiresto build the process relationshipsnum_retries— How many times to retry to jobs once error occursorder— The execution order for this process. The bigger the numberis, the later the process will be executed. Default: 0. Note that the dependent processes will always be executed first. This doesn't work for start processes either, whose orders are determined byPipen.set_starts()output— The output keys for the output channel(the data will be computed)output_data— The output data (to pass to the next processes)plugin_opts— Options for process-level pluginsrequires— The dependency processesscheduler— The scheduler to run the jobsscheduler_opts— The options for the schedulerscript— The script template for the processsubmission_batch— How many jobs to be submited simultaneously.The program entrance for some schedulers may take too much resources when submitting a job or checking the job status. So we may use a smaller number here to limit the simultaneous submissions.template— Define the template engine to use.This could be either a template engine or a dict with keyengineindicating the template engine and the rest the arguments passed to the constructor of thepipen.template.Templateobject. The template engine could be either the name of the engine, currently jinja2 and liquidpy are supported, or a subclass ofpipen.template.Template. You can subclasspipen.template.Templateto use your own template engine.
scrfile— The data loaded byScRepCombiningExpression, saved in RDS or qs/qs2 format.The data is actually generated byscRepertiore::combineExpression(). The data must have both TRA and TRB chains.
outdir— The output directory
comparison(type=auto) — A dict of two groups, with keys as thegroup names and values as the group labels. For example,Or simply a list of two groups, for example,Treg = ["CD4 CTL", "CD4 Naive", "CD4 TCM", "CD4 TEM"] Tconv = "Tconv"["Treg", "Tconv"]when they are both in thegroupcolumn.each(auto) — A column, or a list of columns or a string of columns separated by comma.The columns will be used to split the data into multiple groups and the regression will be applied to each group separately. If not provided, all the cells will be used.group— The key of group in metadata to define the groups tocompare. For example,CellType, which has cell types annotated for each cell in the combined object (immdata + Seurat metadata)target— Which group to use as the target group. The targetgroup will be labeled as 1, and the other group will be labeled as 0 in the regression. If not specified, the first group incomparisonwill be used as the target group.
__init_subclass__()— Do the requirements inferring since we need them to build up theprocess relationship </>from_proc(proc,name,desc,envs,envs_depth,cache,export,error_strategy,num_retries,forks,input_data,order,plugin_opts,requires,scheduler,scheduler_opts,submission_batch)(Type) — Create a subclass of Proc using another Proc subclass or Proc itself</>gc()— GC process for the process to save memory after it's done</>log(level,msg,*args,logger)— Log message for the process</>run()— Init all other properties and jobs</>
pipen.proc.ProcMeta(name, bases, namespace, **kwargs)
Meta class for Proc
__call__(cls,*args,**kwds)(Proc) — Make sure Proc subclasses are singletons</>__instancecheck__(cls,instance)— Override for isinstance(instance, cls).</>__repr__(cls)(str) — Representation for the Proc subclasses</>__subclasscheck__(cls,subclass)— Override for issubclass(subclass, cls).</>register(cls,subclass)— Register a virtual subclass of an ABC.</>
register(cls, subclass)Register a virtual subclass of an ABC.
Returns the subclass, to allow usage as a class decorator.
__instancecheck__(cls, instance)Override for isinstance(instance, cls).
__subclasscheck__(cls, subclass)Override for issubclass(subclass, cls).
__repr__(cls) → strRepresentation for the Proc subclasses
__call__(cls, *args, **kwds)Make sure Proc subclasses are singletons
*args(Any) — and**kwds(Any) — Arguments for the constructor
The Proc instance
from_proc(proc, name=None, desc=None, envs=None, envs_depth=None, cache=None, export=None, error_strategy=None, num_retries=None, forks=None, input_data=None, order=None, plugin_opts=None, requires=None, scheduler=None, scheduler_opts=None, submission_batch=None)
Create a subclass of Proc using another Proc subclass or Proc itself
proc(Type) — The Proc subclassname(str, optional) — The new name of the processdesc(str, optional) — The new description of the processenvs(Mapping, optional) — The arguments of the process, will overwrite parent oneThe items that are specified will be inheritedenvs_depth(int, optional) — How deep to update the envs when subclassed.cache(bool, optional) — Whether we should check the cache for the jobsexport(bool, optional) — When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processeserror_strategy(str, optional) — How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
num_retries(int, optional) — How many times to retry to jobs once error occursforks(int, optional) — New forks for the new processinput_data(Any, optional) — The input data for the process. Only when this processis a start processorder(int, optional) — The order to execute the new processplugin_opts(Mapping, optional) — The new plugin options, unspecified items will beinherited.requires(Sequence, optional) — The required processes for the new processscheduler(str, optional) — The new shedular to run the new processscheduler_opts(Mapping, optional) — The new scheduler options, unspecified items willbe inherited.submission_batch(int, optional) — How many jobs to be submited simultaneously.
The new process class
__init_subclass__()
Do the requirements inferring since we need them to build up theprocess relationship
run()
Init all other properties and jobs
gc()
GC process for the process to save memory after it's done
log(level, msg, *args, logger=<LoggerAdapter pipen.core (WARNING)>)
Log message for the process
level(int | str) — The log level of the recordmsg(str) — The message to log*args— The arguments to format the messagelogger(LoggerAdapter, optional) — The logging logger
biopipen.ns.tcr.TESSA(*args, **kwds) → Proc
Tessa is a Bayesian model to integrate T cell receptor (TCR) sequenceprofiling with transcriptomes of T cells.
Enabled by the recently developed single cell sequencing techniques, which provide both TCR sequences and RNA sequences of each T cell concurrently, Tessa maps the functional landscape of the TCR repertoire, and generates insights into understanding human immune response to diseases. As the first part of tessa, BriseisEncoder is employed prior to the Bayesian algorithm to capture the TCR sequence features and create numerical embeddings. We showed that the reconstructed Atchley Factor matrices and CDR3 sequences, generated through the numerical embeddings, are highly similar to their original counterparts. The CDR3 peptide sequences are constructed via a RandomForest model applied on the reconstructed Atchley Factor matrices.
See https://github.com/jcao89757/TESSA
When finished, two columns will be added to the meta.data of the Seurat object:
TESSA_Cluster: The cluster assignments from TESSA.TESSA_Cluster_Size: The number of cells in each cluster.
These columns can be then used for further downstream analysis to explore the functional landscape of the TCR repertoire.
Reference: - 'Mapping the Functional Landscape of TCR Repertoire.', Zhang, Z., Xiong, D., Wang, X. et al. 2021. link - 'Deep learning-based prediction of the T cell receptor-antigen binding specificity.', Lu, T., Zhang, Z., Zhu, J. et al. 2021. link
cache— Should we detect whether the jobs are cached?desc— The description of the process. Will use the summary fromthe docstring by default.dirsig— When checking the signature for caching, whether should we walkthrough the content of the directory? This is sometimes time-consuming if the directory is big.envs— The arguments that are job-independent, useful for common optionsacross jobs.envs_depth— How deep to update the envs when subclassed.error_strategy— How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
export— When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processesforks— How many jobs to run simultaneously?input— The keys for the input channelinput_data— The input data (will be computed for dependent processes)lang— The language for the script to run. Should be the path to theinterpreter iflangis not in$PATH.name— The name of the process. Will use the class name by default.nexts— Computed fromrequiresto build the process relationshipsnum_retries— How many times to retry to jobs once error occursorder— The execution order for this process. The bigger the numberis, the later the process will be executed. Default: 0. Note that the dependent processes will always be executed first. This doesn't work for start processes either, whose orders are determined byPipen.set_starts()output— The output keys for the output channel(the data will be computed)output_data— The output data (to pass to the next processes)plugin_opts— Options for process-level pluginsrequires— The dependency processesscheduler— The scheduler to run the jobsscheduler_opts— The options for the schedulerscript— The script template for the processsubmission_batch— How many jobs to be submited simultaneously.The program entrance for some schedulers may take too much resources when submitting a job or checking the job status. So we may use a smaller number here to limit the simultaneous submissions.template— Define the template engine to use.This could be either a template engine or a dict with keyengineindicating the template engine and the rest the arguments passed to the constructor of thepipen.template.Templateobject. The template engine could be either the name of the engine, currently jinja2 and liquidpy are supported, or a subclass ofpipen.template.Template. You can subclasspipen.template.Templateto use your own template engine.
screpdata— The data loaded byScRepCombiningExpression, saved in RDS orqs/qs2 format. The data is actually generated byscRepertiore::combineExpression(). The data must have both TRA and TRB chains.
outfile— a qs fileof a Seurat object, withTESSA_ClusterandTESSA_Cluster_Sizeadded to themeta.data
assay— Which assay to use to extract the expression matrix.Only works ifin.srtobjis an RDS file of a Seurat object. By default, ifSCTransformis performed,SCTwill be used.max_iter(type=int) — The maximum number of iterations for MCMC.predefined_b(flag) — Whether use the predefinedbor not.Please check the paper of tessa for more details about the b vector. If True, the tessa will not update b in the MCMC iterations.python— The path of python withTESSA's dependencies installedsave_tessa(flag) — Save tessa detailed results to seurat object?It will be saved tosobj@misc$tessa.within_sample(flag) — Whether the TCR networks are constructed onlywithin TCRs from the same sample/patient (True) or with all the TCRs in the meta data matrix (False).
__init_subclass__()— Do the requirements inferring since we need them to build up theprocess relationship </>from_proc(proc,name,desc,envs,envs_depth,cache,export,error_strategy,num_retries,forks,input_data,order,plugin_opts,requires,scheduler,scheduler_opts,submission_batch)(Type) — Create a subclass of Proc using another Proc subclass or Proc itself</>gc()— GC process for the process to save memory after it's done</>log(level,msg,*args,logger)— Log message for the process</>run()— Init all other properties and jobs</>
pipen.proc.ProcMeta(name, bases, namespace, **kwargs)
Meta class for Proc
__call__(cls,*args,**kwds)(Proc) — Make sure Proc subclasses are singletons</>__instancecheck__(cls,instance)— Override for isinstance(instance, cls).</>__repr__(cls)(str) — Representation for the Proc subclasses</>__subclasscheck__(cls,subclass)— Override for issubclass(subclass, cls).</>register(cls,subclass)— Register a virtual subclass of an ABC.</>
register(cls, subclass)Register a virtual subclass of an ABC.
Returns the subclass, to allow usage as a class decorator.
__instancecheck__(cls, instance)Override for isinstance(instance, cls).
__subclasscheck__(cls, subclass)Override for issubclass(subclass, cls).
__repr__(cls) → strRepresentation for the Proc subclasses
__call__(cls, *args, **kwds)Make sure Proc subclasses are singletons
*args(Any) — and**kwds(Any) — Arguments for the constructor
The Proc instance
from_proc(proc, name=None, desc=None, envs=None, envs_depth=None, cache=None, export=None, error_strategy=None, num_retries=None, forks=None, input_data=None, order=None, plugin_opts=None, requires=None, scheduler=None, scheduler_opts=None, submission_batch=None)
Create a subclass of Proc using another Proc subclass or Proc itself
proc(Type) — The Proc subclassname(str, optional) — The new name of the processdesc(str, optional) — The new description of the processenvs(Mapping, optional) — The arguments of the process, will overwrite parent oneThe items that are specified will be inheritedenvs_depth(int, optional) — How deep to update the envs when subclassed.cache(bool, optional) — Whether we should check the cache for the jobsexport(bool, optional) — When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processeserror_strategy(str, optional) — How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
num_retries(int, optional) — How many times to retry to jobs once error occursforks(int, optional) — New forks for the new processinput_data(Any, optional) — The input data for the process. Only when this processis a start processorder(int, optional) — The order to execute the new processplugin_opts(Mapping, optional) — The new plugin options, unspecified items will beinherited.requires(Sequence, optional) — The required processes for the new processscheduler(str, optional) — The new shedular to run the new processscheduler_opts(Mapping, optional) — The new scheduler options, unspecified items willbe inherited.submission_batch(int, optional) — How many jobs to be submited simultaneously.
The new process class
__init_subclass__()
Do the requirements inferring since we need them to build up theprocess relationship
run()
Init all other properties and jobs
gc()
GC process for the process to save memory after it's done
log(level, msg, *args, logger=<LoggerAdapter pipen.core (WARNING)>)
Log message for the process
level(int | str) — The log level of the recordmsg(str) — The message to log*args— The arguments to format the messagelogger(LoggerAdapter, optional) — The logging logger
biopipen.ns.tcr.TCRDock(*args, **kwds) → Proc
Using TCRDock to predict the structure of MHC-peptide-TCR complexes
cache— Should we detect whether the jobs are cached?desc— The description of the process. Will use the summary fromthe docstring by default.dirsig— When checking the signature for caching, whether should we walkthrough the content of the directory? This is sometimes time-consuming if the directory is big.envs— The arguments that are job-independent, useful for common optionsacross jobs.envs_depth— How deep to update the envs when subclassed.error_strategy— How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
export— When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processesforks— How many jobs to run simultaneously?input— The keys for the input channelinput_data— The input data (will be computed for dependent processes)lang— The language for the script to run. Should be the path to theinterpreter iflangis not in$PATH.name— The name of the process. Will use the class name by default.nexts— Computed fromrequiresto build the process relationshipsnum_retries— How many times to retry to jobs once error occursorder— The execution order for this process. The bigger the numberis, the later the process will be executed. Default: 0. Note that the dependent processes will always be executed first. This doesn't work for start processes either, whose orders are determined byPipen.set_starts()output— The output keys for the output channel(the data will be computed)output_data— The output data (to pass to the next processes)plugin_opts— Options for process-level pluginsrequires— The dependency processesscheduler— The scheduler to run the jobsscheduler_opts— The options for the schedulerscript— The script template for the processsubmission_batch— How many jobs to be submited simultaneously.The program entrance for some schedulers may take too much resources when submitting a job or checking the job status. So we may use a smaller number here to limit the simultaneous submissions.template— Define the template engine to use.This could be either a template engine or a dict with keyengineindicating the template engine and the rest the arguments passed to the constructor of thepipen.template.Templateobject. The template engine could be either the name of the engine, currently jinja2 and liquidpy are supported, or a subclass ofpipen.template.Template. You can subclasspipen.template.Templateto use your own template engine.
configfile— The config file for TCRDockIt's should be a toml file with the keys listed inenvs, includingorganism,mhc_class,mhc,peptide,va,ja,vb,jb,cdr3a, andcdr3b. The values will overwrite the values inenvs.
outdir— The output directory containing the results
cdr3a— The CDR3 alpha sequencecdr3b— The CDR3 beta sequencedata_dir— The data directory that contains the model files.The model files should be in theparamssubdirectory.ja— The J alpha genejb— The J beta genemhc— The MHC allele, e.g.,A*02:01mhc_class(type=int) — The MHC class, either1or2model_file— The model file to use.If provided as a relative path, it should be relative to the<envs.data_dir>/params/, otherwise, it should be the full path.model_name— The model name to useorganism— The organism of the TCR, peptide and MHCpeptide— The peptide sequencepython— The path of python with dependencies fortcrdockinstalled.If not provided,TCRDock.langwill be used (the same interpreter used for the wrapper script). It could also be a list to specify, for example, a python in a conda environment (e.g.,["conda", "run", "-n", "myenv", "python"]).tcrdock— The path to thetcrdocksource code repo.You need to clone the source code from the github repository. https://github.com/phbradley/TCRdock at revision c5a7af42eeb0c2a4492a4d4fe803f1f9aafb6193 at main branch. You also have to rundownload_blast.pyafter cloning to download the blast database in the directory. If not provided, we will clone the source code to theenvs.tmpdirdirectory and run thedownload_blast.pyscript.tmpdir— The temporary directory used to clone thetcrdocksource code ifenvs.tcrdockis not provided.va— The V alpha genevb— The V beta gene
__init_subclass__()— Do the requirements inferring since we need them to build up theprocess relationship </>from_proc(proc,name,desc,envs,envs_depth,cache,export,error_strategy,num_retries,forks,input_data,order,plugin_opts,requires,scheduler,scheduler_opts,submission_batch)(Type) — Create a subclass of Proc using another Proc subclass or Proc itself</>gc()— GC process for the process to save memory after it's done</>log(level,msg,*args,logger)— Log message for the process</>run()— Init all other properties and jobs</>
pipen.proc.ProcMeta(name, bases, namespace, **kwargs)
Meta class for Proc
__call__(cls,*args,**kwds)(Proc) — Make sure Proc subclasses are singletons</>__instancecheck__(cls,instance)— Override for isinstance(instance, cls).</>__repr__(cls)(str) — Representation for the Proc subclasses</>__subclasscheck__(cls,subclass)— Override for issubclass(subclass, cls).</>register(cls,subclass)— Register a virtual subclass of an ABC.</>
register(cls, subclass)Register a virtual subclass of an ABC.
Returns the subclass, to allow usage as a class decorator.
__instancecheck__(cls, instance)Override for isinstance(instance, cls).
__subclasscheck__(cls, subclass)Override for issubclass(subclass, cls).
__repr__(cls) → strRepresentation for the Proc subclasses
__call__(cls, *args, **kwds)Make sure Proc subclasses are singletons
*args(Any) — and**kwds(Any) — Arguments for the constructor
The Proc instance
from_proc(proc, name=None, desc=None, envs=None, envs_depth=None, cache=None, export=None, error_strategy=None, num_retries=None, forks=None, input_data=None, order=None, plugin_opts=None, requires=None, scheduler=None, scheduler_opts=None, submission_batch=None)
Create a subclass of Proc using another Proc subclass or Proc itself
proc(Type) — The Proc subclassname(str, optional) — The new name of the processdesc(str, optional) — The new description of the processenvs(Mapping, optional) — The arguments of the process, will overwrite parent oneThe items that are specified will be inheritedenvs_depth(int, optional) — How deep to update the envs when subclassed.cache(bool, optional) — Whether we should check the cache for the jobsexport(bool, optional) — When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processeserror_strategy(str, optional) — How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
num_retries(int, optional) — How many times to retry to jobs once error occursforks(int, optional) — New forks for the new processinput_data(Any, optional) — The input data for the process. Only when this processis a start processorder(int, optional) — The order to execute the new processplugin_opts(Mapping, optional) — The new plugin options, unspecified items will beinherited.requires(Sequence, optional) — The required processes for the new processscheduler(str, optional) — The new shedular to run the new processscheduler_opts(Mapping, optional) — The new scheduler options, unspecified items willbe inherited.submission_batch(int, optional) — How many jobs to be submited simultaneously.
The new process class
__init_subclass__()
Do the requirements inferring since we need them to build up theprocess relationship
run()
Init all other properties and jobs
gc()
GC process for the process to save memory after it's done
log(level, msg, *args, logger=<LoggerAdapter pipen.core (WARNING)>)
Log message for the process
level(int | str) — The log level of the recordmsg(str) — The message to log*args— The arguments to format the messagelogger(LoggerAdapter, optional) — The logging logger
biopipen.ns.tcr.ScRepLoading(*args, **kwds) → Proc
Load the single cell TCR/BCR data into a scRepertoire compatible object
This process loads the single cell TCR/BCR data into a scRepertoire
(>= v2.0.8, < v2.3.2) compatible object. Later, scRepertoire::combineExpression
can be used to combine the expression data with the TCR/BCR data.
For the data path specified at TCRData/BCRData in the input file
(in.metafile), will be used to find the TCR/BCR data files and
scRepertoire::loadContigs() will be used to load the data.
A directory can be specified in TCRData/BCRData, then
scRepertoire::loadContigs() will be used directly to load the data from the
directory. Otherwise if a file is specified, it will be symbolically linked to
a directory for scRepertoire::loadContigs() to load.
Note that when the file name can not be recognized by scRepertoire::loadContigs(),
envs.format must be set for the correct format of the data.
cache— Should we detect whether the jobs are cached?desc— The description of the process. Will use the summary fromthe docstring by default.dirsig— When checking the signature for caching, whether should we walkthrough the content of the directory? This is sometimes time-consuming if the directory is big.envs— The arguments that are job-independent, useful for common optionsacross jobs.envs_depth— How deep to update the envs when subclassed.error_strategy— How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
export— When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processesforks— How many jobs to run simultaneously?input— The keys for the input channelinput_data— The input data (will be computed for dependent processes)lang— The language for the script to run. Should be the path to theinterpreter iflangis not in$PATH.name— The name of the process. Will use the class name by default.nexts— Computed fromrequiresto build the process relationshipsnum_retries— How many times to retry to jobs once error occursorder— The execution order for this process. The bigger the numberis, the later the process will be executed. Default: 0. Note that the dependent processes will always be executed first. This doesn't work for start processes either, whose orders are determined byPipen.set_starts()output— The output keys for the output channel(the data will be computed)output_data— The output data (to pass to the next processes)plugin_opts— Options for process-level pluginsrequires— The dependency processesscheduler— The scheduler to run the jobsscheduler_opts— The options for the schedulerscript— The script template for the processsubmission_batch— How many jobs to be submited simultaneously.The program entrance for some schedulers may take too much resources when submitting a job or checking the job status. So we may use a smaller number here to limit the simultaneous submissions.template— Define the template engine to use.This could be either a template engine or a dict with keyengineindicating the template engine and the rest the arguments passed to the constructor of thepipen.template.Templateobject. The template engine could be either the name of the engine, currently jinja2 and liquidpy are supported, or a subclass ofpipen.template.Template. You can subclasspipen.template.Templateto use your own template engine.
metafile— The meta data of the samplesA tab-delimited file Two columns are required:- *
Sampleto specify the sample names. - *
TCRData/BCRDatato assign the path of the data to the samples,
- *
outfile— ThescRepertoirecompatible object in qs/qs2 format
combineBCR(type=json) — The extra arguments forscRepertoire::combineBCRfunction. See also https://www.borch.dev/uploads/screpertoire/reference/combinebcrcombineTCR(type=json) — The extra arguments forscRepertoire::combineTCRfunction. See also https://www.borch.dev/uploads/screpertoire/reference/combinetcrexclude(auto) — The columns to exclude from the metadata to add to the object.A list of column names to exclude or a string with column names separated by,. By default,BCRData,TCRDataandRNADatawill be excluded.format(choice) — The format of the TCR/BCR data files.- - 10X: 10X Genomics data, which is usually in a directory with
filtered_contig_annotations.csvfile. - - AIRR: AIRR format, which is usually in a file with
airr_rearrangement.tsvfile. - - BD: Becton Dickinson data, which is usually in a file with
Contigs_AIRR.tsvfile. - - Dandelion: Dandelion data, which is usually in a file with
all_contig_dandelion.tsvfile. - - Immcantation: Immcantation data, which is usually in a file with
data.tsvfile. - - JSON: JSON format, which is usually in a file with
.jsonextension. - - ParseBio: ParseBio data, which is usually in a file with
barcode_report.tsvfile. - - MiXCR: MiXCR data, which is usually in a file with
clones.tsvfile. - - Omniscope: Omniscope data, which is usually in a file with
.csv
extension. - - TRUST4: TRUST4 data, which is usually in a file with
barcode_report.tsvfile. - - WAT3R: WAT3R data, which is usually in a file with
barcode_results.csvfile.
scRepertoire::loadContigs().- - 10X: 10X Genomics data, which is usually in a directory with
tmpdir— The temporary directory to store the symbolic links to theTCR/BCR data files.type(choice) — The type of the data to load.- - TCR: T cell receptor data
- - BCR: B cell receptor data
- - auto: Automatically detect the type from the metadata.
Ifautois selected, the type will be determined by the presence of
TCRDataorBCRDatacolumns in the metadata. If both columns are
present,TCRwill be selected by default.
__init_subclass__()— Do the requirements inferring since we need them to build up theprocess relationship </>from_proc(proc,name,desc,envs,envs_depth,cache,export,error_strategy,num_retries,forks,input_data,order,plugin_opts,requires,scheduler,scheduler_opts,submission_batch)(Type) — Create a subclass of Proc using another Proc subclass or Proc itself</>gc()— GC process for the process to save memory after it's done</>log(level,msg,*args,logger)— Log message for the process</>run()— Init all other properties and jobs</>
pipen.proc.ProcMeta(name, bases, namespace, **kwargs)
Meta class for Proc
__call__(cls,*args,**kwds)(Proc) — Make sure Proc subclasses are singletons</>__instancecheck__(cls,instance)— Override for isinstance(instance, cls).</>__repr__(cls)(str) — Representation for the Proc subclasses</>__subclasscheck__(cls,subclass)— Override for issubclass(subclass, cls).</>register(cls,subclass)— Register a virtual subclass of an ABC.</>
register(cls, subclass)Register a virtual subclass of an ABC.
Returns the subclass, to allow usage as a class decorator.
__instancecheck__(cls, instance)Override for isinstance(instance, cls).
__subclasscheck__(cls, subclass)Override for issubclass(subclass, cls).
__repr__(cls) → strRepresentation for the Proc subclasses
__call__(cls, *args, **kwds)Make sure Proc subclasses are singletons
*args(Any) — and**kwds(Any) — Arguments for the constructor
The Proc instance
from_proc(proc, name=None, desc=None, envs=None, envs_depth=None, cache=None, export=None, error_strategy=None, num_retries=None, forks=None, input_data=None, order=None, plugin_opts=None, requires=None, scheduler=None, scheduler_opts=None, submission_batch=None)
Create a subclass of Proc using another Proc subclass or Proc itself
proc(Type) — The Proc subclassname(str, optional) — The new name of the processdesc(str, optional) — The new description of the processenvs(Mapping, optional) — The arguments of the process, will overwrite parent oneThe items that are specified will be inheritedenvs_depth(int, optional) — How deep to update the envs when subclassed.cache(bool, optional) — Whether we should check the cache for the jobsexport(bool, optional) — When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processeserror_strategy(str, optional) — How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
num_retries(int, optional) — How many times to retry to jobs once error occursforks(int, optional) — New forks for the new processinput_data(Any, optional) — The input data for the process. Only when this processis a start processorder(int, optional) — The order to execute the new processplugin_opts(Mapping, optional) — The new plugin options, unspecified items will beinherited.requires(Sequence, optional) — The required processes for the new processscheduler(str, optional) — The new shedular to run the new processscheduler_opts(Mapping, optional) — The new scheduler options, unspecified items willbe inherited.submission_batch(int, optional) — How many jobs to be submited simultaneously.
The new process class
__init_subclass__()
Do the requirements inferring since we need them to build up theprocess relationship
run()
Init all other properties and jobs
gc()
GC process for the process to save memory after it's done
log(level, msg, *args, logger=<LoggerAdapter pipen.core (WARNING)>)
Log message for the process
level(int | str) — The log level of the recordmsg(str) — The message to log*args— The arguments to format the messagelogger(LoggerAdapter, optional) — The logging logger
biopipen.ns.tcr.ScRepCombiningExpression(*args, **kwds) → Proc
Combine the scTCR/BCR data with the expression data
This process combines the scTCR/BCR data with the expression data using
scRepertoire::combineExpression function. The expression data should be
in Seurat format. The scRepertoire object should be a combined contig
object, usually generated by scRepertoire::combineTCR or
scRepertoire::combineBCR.
See also: https://www.borch.dev/uploads/screpertoire/reference/combineexpression.
cache— Should we detect whether the jobs are cached?desc— The description of the process. Will use the summary fromthe docstring by default.dirsig— When checking the signature for caching, whether should we walkthrough the content of the directory? This is sometimes time-consuming if the directory is big.envs— The arguments that are job-independent, useful for common optionsacross jobs.envs_depth— How deep to update the envs when subclassed.error_strategy— How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
export— When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processesforks— How many jobs to run simultaneously?input— The keys for the input channelinput_data— The input data (will be computed for dependent processes)lang— The language for the script to run. Should be the path to theinterpreter iflangis not in$PATH.name— The name of the process. Will use the class name by default.nexts— Computed fromrequiresto build the process relationshipsnum_retries— How many times to retry to jobs once error occursorder— The execution order for this process. The bigger the numberis, the later the process will be executed. Default: 0. Note that the dependent processes will always be executed first. This doesn't work for start processes either, whose orders are determined byPipen.set_starts()output— The output keys for the output channel(the data will be computed)output_data— The output data (to pass to the next processes)plugin_opts— Options for process-level pluginsrequires— The dependency processesscheduler— The scheduler to run the jobsscheduler_opts— The options for the schedulerscript— The script template for the processsubmission_batch— How many jobs to be submited simultaneously.The program entrance for some schedulers may take too much resources when submitting a job or checking the job status. So we may use a smaller number here to limit the simultaneous submissions.template— Define the template engine to use.This could be either a template engine or a dict with keyengineindicating the template engine and the rest the arguments passed to the constructor of thepipen.template.Templateobject. The template engine could be either the name of the engine, currently jinja2 and liquidpy are supported, or a subclass ofpipen.template.Template. You can subclasspipen.template.Templateto use your own template engine.
screpfile— ThescRepertoireobject in RDS/qs formatsrtobj— TheSeuratobject, saved in RDS/qs format
outfile— TheSeuratobject with the TCR/BCR data combinedIn addition to the meta columns added byscRepertoire::combineExpression(), a new columnVDJ_Presencewill be added to the metadata. It indicates whether the cell has a TCR/BCR sequence or not. The value isTRUEif the cell has a TCR/BCR sequence, andFALSEotherwise.
addLabel(flag) — This will add a label to the frequency header, allowing theuser to try multiple group_by variables or recalculate frequencies after subsetting the data.chain— indicate if both or a specific chain should be usede.g. "both", "TRA", "TRG", "IGH", "IGL".cloneCall— How to call the clone - VDJC gene (gene), CDR3 nucleotide (nt),CDR3 amino acid (aa), VDJC gene + CDR3 nucleotide (strict) or a custom variable in the data.cloneSize(type=json) — The bins for the grouping based on proportion orfrequency. If proportion is FALSE and the cloneSizes are not set high enough based on frequency, the upper limit of cloneSizes will be automatically updated.filterNA(flag) — Method to subset Seurat/SCE object of barcodes without cloneinformationgroup_by— The column label in the combined clones in which clone frequency willbe calculated. NULL or "none" will keep the format of input.data.proportion(flag) — Whether to proportion (TRUE) or total frequency (FALSE) ofthe clone based on the group_by variable.
__init_subclass__()— Do the requirements inferring since we need them to build up theprocess relationship </>from_proc(proc,name,desc,envs,envs_depth,cache,export,error_strategy,num_retries,forks,input_data,order,plugin_opts,requires,scheduler,scheduler_opts,submission_batch)(Type) — Create a subclass of Proc using another Proc subclass or Proc itself</>gc()— GC process for the process to save memory after it's done</>log(level,msg,*args,logger)— Log message for the process</>run()— Init all other properties and jobs</>
pipen.proc.ProcMeta(name, bases, namespace, **kwargs)
Meta class for Proc
__call__(cls,*args,**kwds)(Proc) — Make sure Proc subclasses are singletons</>__instancecheck__(cls,instance)— Override for isinstance(instance, cls).</>__repr__(cls)(str) — Representation for the Proc subclasses</>__subclasscheck__(cls,subclass)— Override for issubclass(subclass, cls).</>register(cls,subclass)— Register a virtual subclass of an ABC.</>
register(cls, subclass)Register a virtual subclass of an ABC.
Returns the subclass, to allow usage as a class decorator.
__instancecheck__(cls, instance)Override for isinstance(instance, cls).
__subclasscheck__(cls, subclass)Override for issubclass(subclass, cls).
__repr__(cls) → strRepresentation for the Proc subclasses
__call__(cls, *args, **kwds)Make sure Proc subclasses are singletons
*args(Any) — and**kwds(Any) — Arguments for the constructor
The Proc instance
from_proc(proc, name=None, desc=None, envs=None, envs_depth=None, cache=None, export=None, error_strategy=None, num_retries=None, forks=None, input_data=None, order=None, plugin_opts=None, requires=None, scheduler=None, scheduler_opts=None, submission_batch=None)
Create a subclass of Proc using another Proc subclass or Proc itself
proc(Type) — The Proc subclassname(str, optional) — The new name of the processdesc(str, optional) — The new description of the processenvs(Mapping, optional) — The arguments of the process, will overwrite parent oneThe items that are specified will be inheritedenvs_depth(int, optional) — How deep to update the envs when subclassed.cache(bool, optional) — Whether we should check the cache for the jobsexport(bool, optional) — When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processeserror_strategy(str, optional) — How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
num_retries(int, optional) — How many times to retry to jobs once error occursforks(int, optional) — New forks for the new processinput_data(Any, optional) — The input data for the process. Only when this processis a start processorder(int, optional) — The order to execute the new processplugin_opts(Mapping, optional) — The new plugin options, unspecified items will beinherited.requires(Sequence, optional) — The required processes for the new processscheduler(str, optional) — The new shedular to run the new processscheduler_opts(Mapping, optional) — The new scheduler options, unspecified items willbe inherited.submission_batch(int, optional) — How many jobs to be submited simultaneously.
The new process class
__init_subclass__()
Do the requirements inferring since we need them to build up theprocess relationship
run()
Init all other properties and jobs
gc()
GC process for the process to save memory after it's done
log(level, msg, *args, logger=<LoggerAdapter pipen.core (WARNING)>)
Log message for the process
level(int | str) — The log level of the recordmsg(str) — The message to log*args— The arguments to format the messagelogger(LoggerAdapter, optional) — The logging logger
biopipen.ns.tcr.ClonalStats(*args, **kwds) → Proc
Visualize the clonal information.
Using scplotter to visualize the clonal
information.
cache— Should we detect whether the jobs are cached?desc— The description of the process. Will use the summary fromthe docstring by default.dirsig— When checking the signature for caching, whether should we walkthrough the content of the directory? This is sometimes time-consuming if the directory is big.envs— The arguments that are job-independent, useful for common optionsacross jobs.envs_depth— How deep to update the envs when subclassed.error_strategy— How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
export— When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processesforks— How many jobs to run simultaneously?input— The keys for the input channelinput_data— The input data (will be computed for dependent processes)lang— The language for the script to run. Should be the path to theinterpreter iflangis not in$PATH.name— The name of the process. Will use the class name by default.nexts— Computed fromrequiresto build the process relationshipsnum_retries— How many times to retry to jobs once error occursorder— The execution order for this process. The bigger the numberis, the later the process will be executed. Default: 0. Note that the dependent processes will always be executed first. This doesn't work for start processes either, whose orders are determined byPipen.set_starts()output— The output keys for the output channel(the data will be computed)output_data— The output data (to pass to the next processes)plugin_opts— Options for process-level pluginsrequires— The dependency processesscheduler— The scheduler to run the jobsscheduler_opts— The options for the schedulerscript— The script template for the processsubmission_batch— How many jobs to be submited simultaneously.The program entrance for some schedulers may take too much resources when submitting a job or checking the job status. So we may use a smaller number here to limit the simultaneous submissions.template— Define the template engine to use.This could be either a template engine or a dict with keyengineindicating the template engine and the rest the arguments passed to the constructor of thepipen.template.Templateobject. The template engine could be either the name of the engine, currently jinja2 and liquidpy are supported, or a subclass ofpipen.template.Template. You can subclasspipen.template.Templateto use your own template engine.
Clonal Volume
[ClonalStats.envs.cases."Clonal Volume"]
viz_type = "volume"
x_text_angle = 45
 {: width="80%"}
{: width="80%"}
Clonal Volume by Diagnosis
[ClonalStats.envs.cases."Clonal Volume by Diagnosis"]
viz_type = "volume"
x = "seurat_clusters"
group_by = "Diagnosis"
comparisons = true
 {: width="80%"}
{: width="80%"}
Clonal Abundance
[ClonalStats.envs.cases."Clonal Abundance"]
viz_type = "abundance"
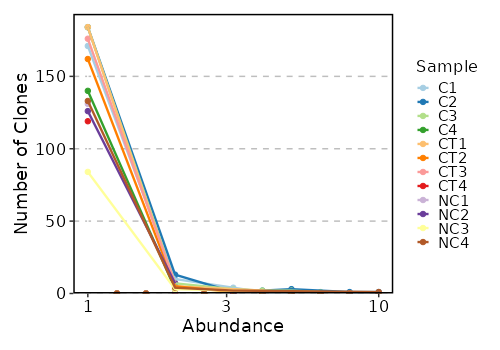 {: width="80%"}
{: width="80%"}
Clonal Abundance Density
[ClonalStats.envs.cases."Clonal Abundance Density"]
viz_type = "abundance"
plot_type = "density"
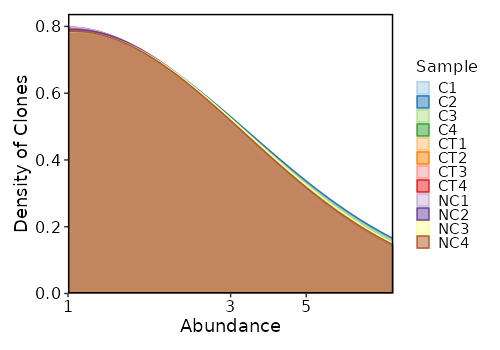 {: width="80%"}
{: width="80%"}
CDR3 Length
[ClonalStats.envs.cases."CDR3 Length"]
viz_type = "length"
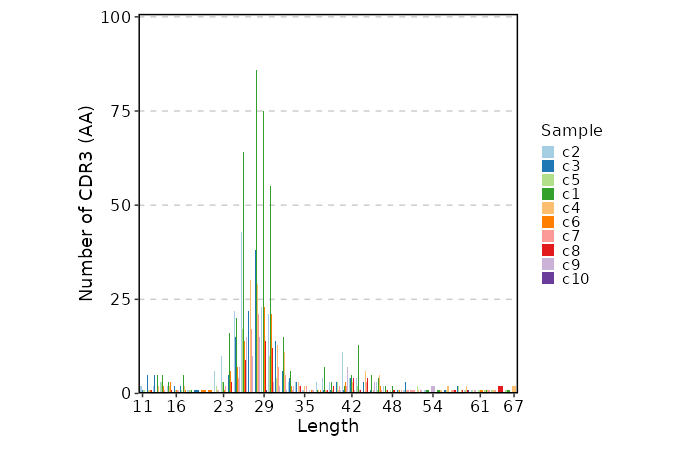 {: width="80%"}
{: width="80%"}
CDR3 Length (Beta Chain)
[ClonalStats.envs.cases."CDR3 Length (Beta Chain)"]
viz_type = "length"
chain = "TRB"
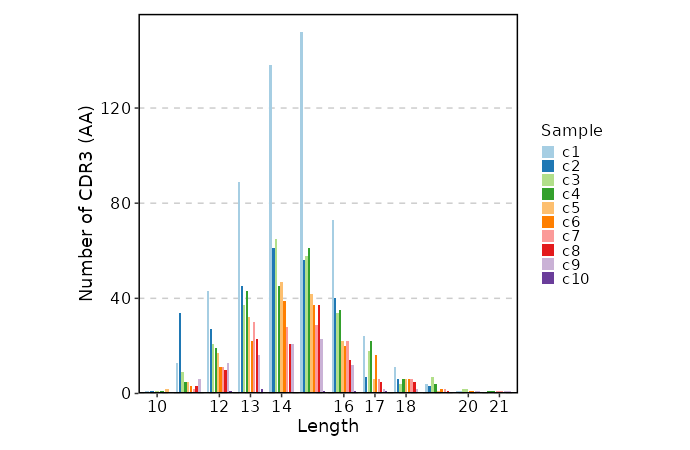 {: width="80%"}
{: width="80%"}
Clonal Residency
[ClonalStats.envs.cases."Clonal Residency"]
viz_type = "residency"
group_by = "Diagnosis"
chain = "TRB"
clone_call = "gene"
groups = ["Colitis", "NoColitis"]
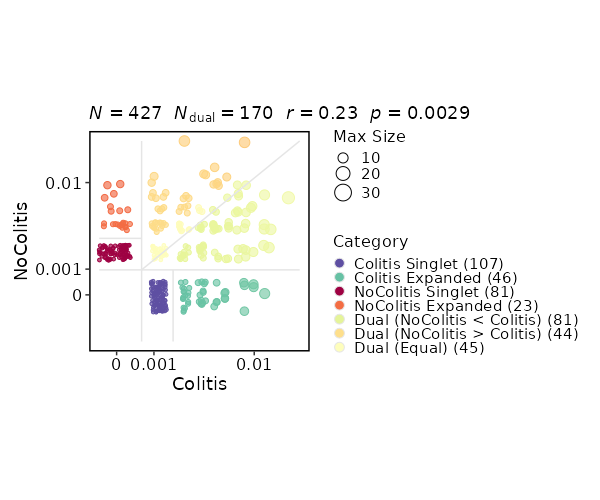 {: width="80%"}
{: width="80%"}
Clonal Residency (UpSet Plot)
[ClonalStats.envs.cases."Clonal Residency (UpSet Plot)"]
viz_type = "residency"
plot_type = "upset"
group_by = "Diagnosis"
chain = "TRB"
clone_call = "gene"
groups = ["Colitis", "NoColitis"]
devpars = {width = 800}
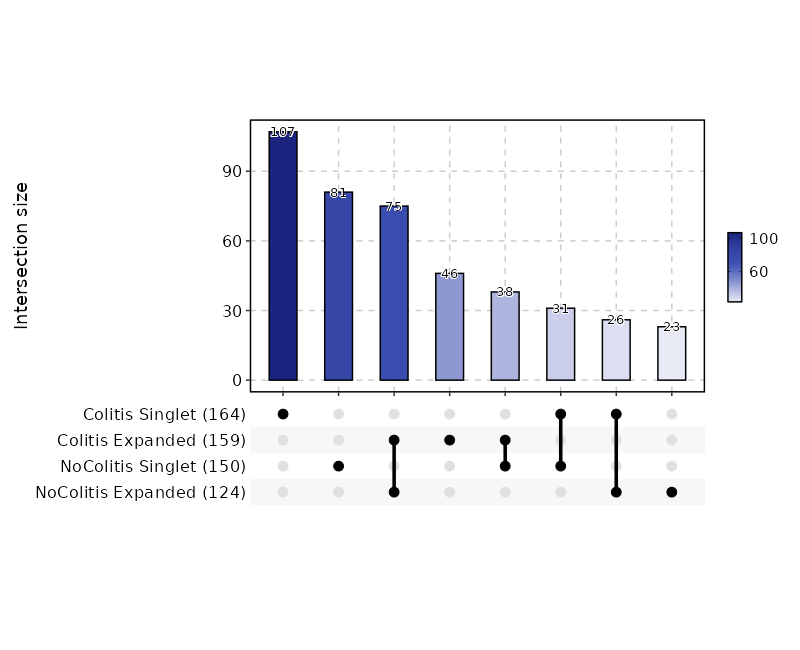 {: width="80%"}
{: width="80%"}
Clonal Statistics with Expanded Clones
[ClonalStats.envs.cases."Clonal Statistics with Expanded Clones"]
viz_type = "stat"
plot_type = "pies"
group_by = "Diagnosis"
groups = ["Colitis", "NoColitis"]
clones = {"Expanded Clones In Colitis" = "sel(Colitis > 2)", "Expanded Clones In NoColitis" = "sel(NoColitis > 2)"}
subgroup_by = "seurat_clusters"
pie_size = "sqrt"
show_row_names = true
show_column_names = true
devpars = {width = 720}
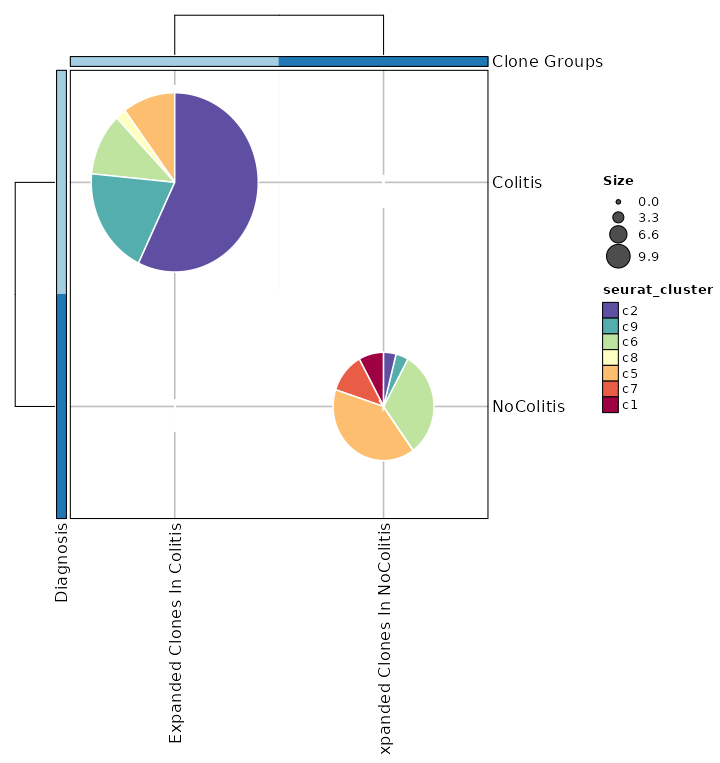 {: width="80%"}
{: width="80%"}
Hyperexpanded Clonal Dynamics
[ClonalStats.envs.cases."Hyperexpanded Clonal Dynamics"]
viz_type = "stat"
plot_type = "sankey"
group_by = "Diagnosis"
chain = "TRB"
groups = ["Colitis", "NoColitis"]
clones = {"Hyper-Expanded Clones In Colitis" = "sel(Colitis > 5)", "Hyper-Expanded Clones In NoColitis" = "sel(NoColitis > 5)"}
devpars = {width = 800}
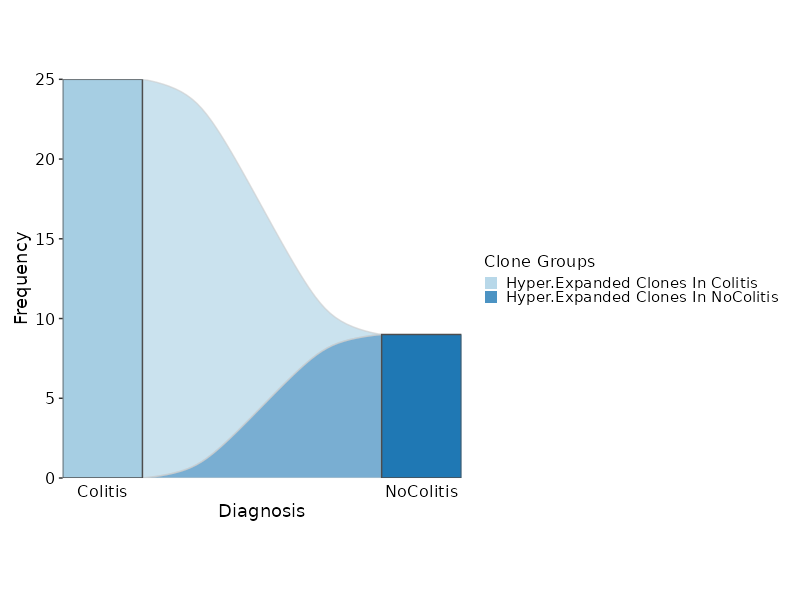 {: width="80%"}
{: width="80%"}
Clonal Composition
[ClonalStats.envs.cases."Clonal Composition"]
viz_type = "composition"
x_text_angle = 45
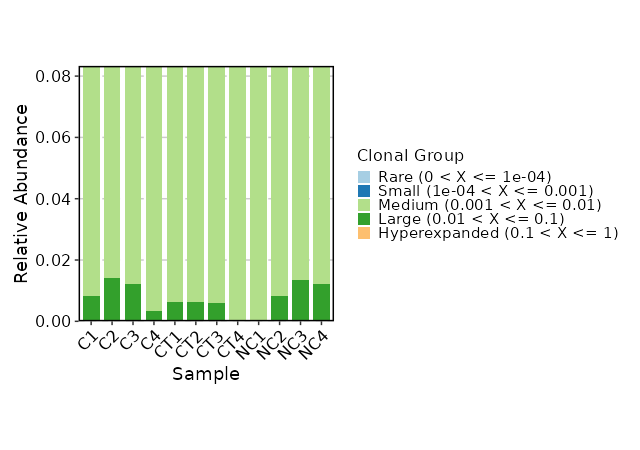 {: width="80%"}
{: width="80%"}
Clonal Overlapping
viz_type = "overlap"
chain = "TRB"
clone_call = "gene"
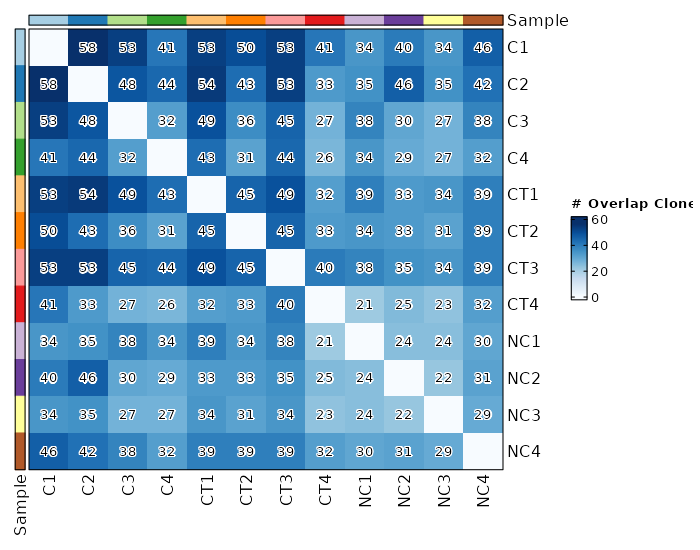 {: width="80%"}
{: width="80%"}
Clonal Diversity
[ClonalStats.envs.cases."Clonal Diversity"]
# method = "shannon" # default
viz_type = "diversity"
x_text_angle = 45
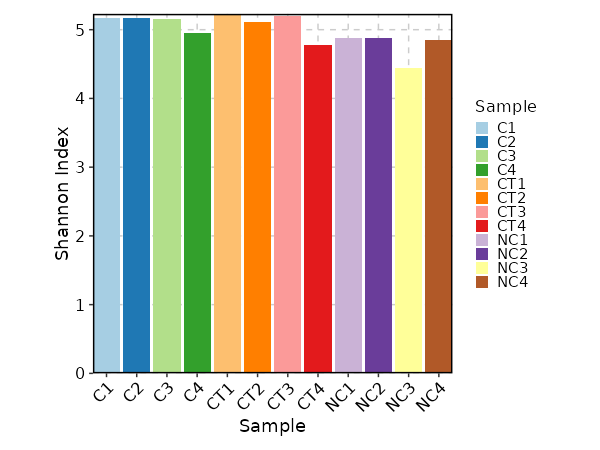 {: width="80%"}
{: width="80%"}
Clonal Diversity (gini.coeff, by Diagnosis)
[ClonalStats.envs.cases."Clonal Diversity (gini.coeff, by Diagnosis)"]
method = "gini.coeff"
viz_type = "diversity"
plot_type = "box"
group_by = "Diagnosis"
comparisons = true
devpars = {height = 600, width = 600}
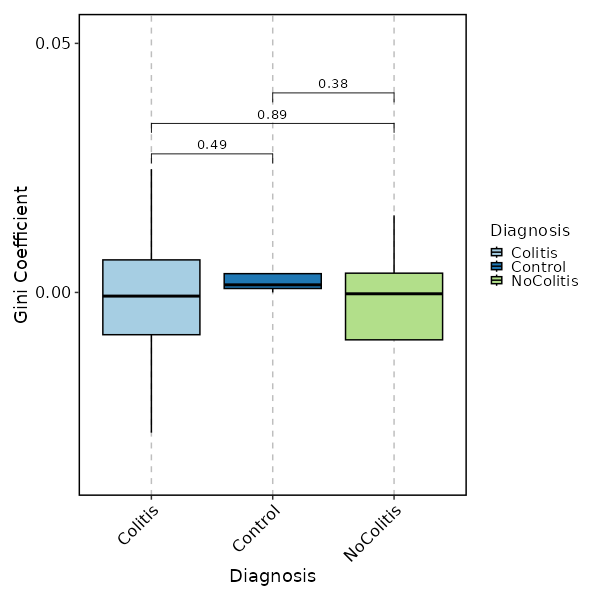 {: width="80%"}
{: width="80%"}
Gene Usage Frequency
[ClonalStats.envs.cases."Gene Usage Frequency"]
viz_type = "geneusage"
devpars = {width = 1200}
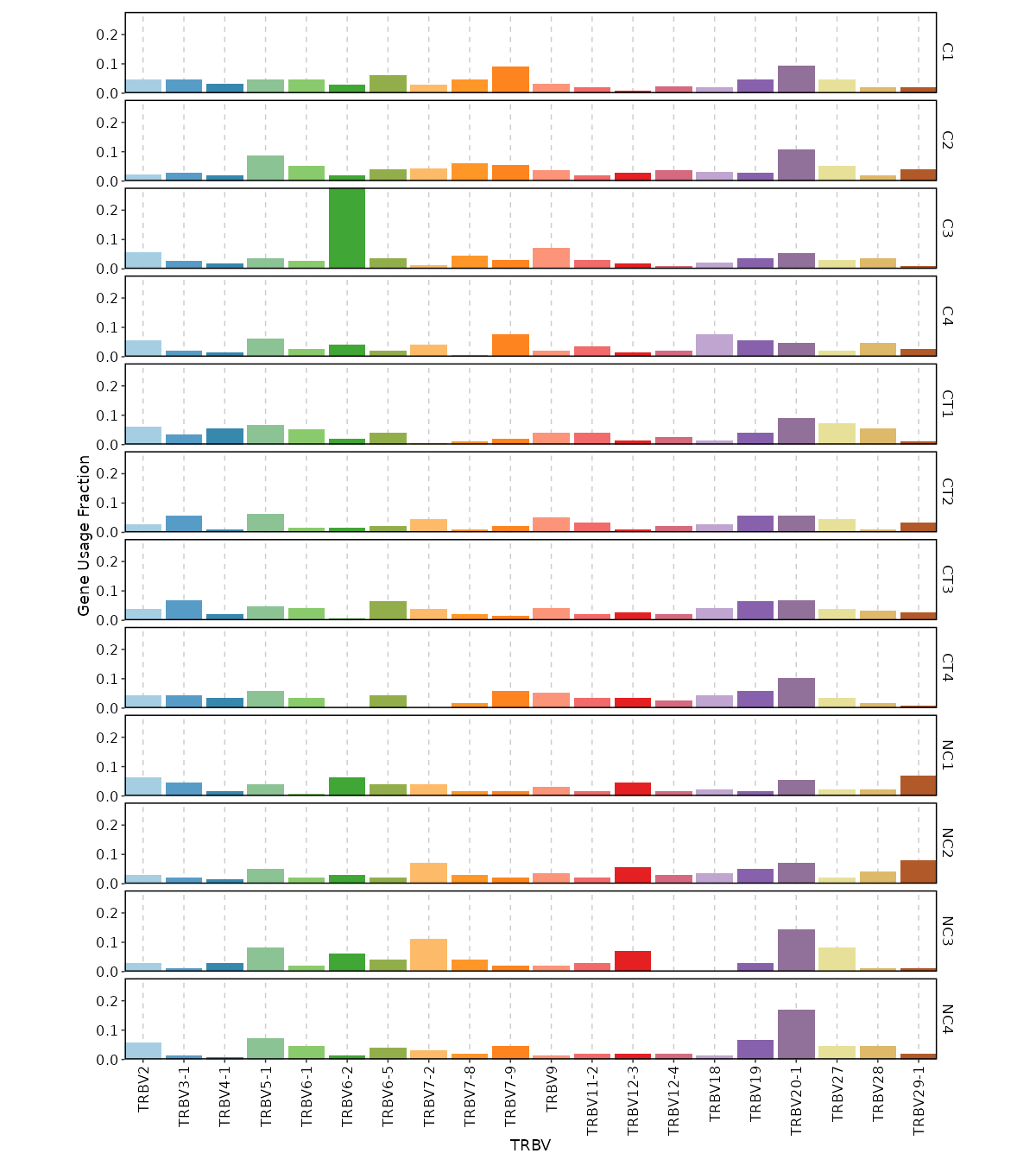 {: width="80%"}
{: width="80%"}
Positional amino acid frequency
[ClonalStats.envs.cases."Positional amino acid frequency"]
viz_type = "positional"
# method = "AA" # default
devpars = {width = 1600}
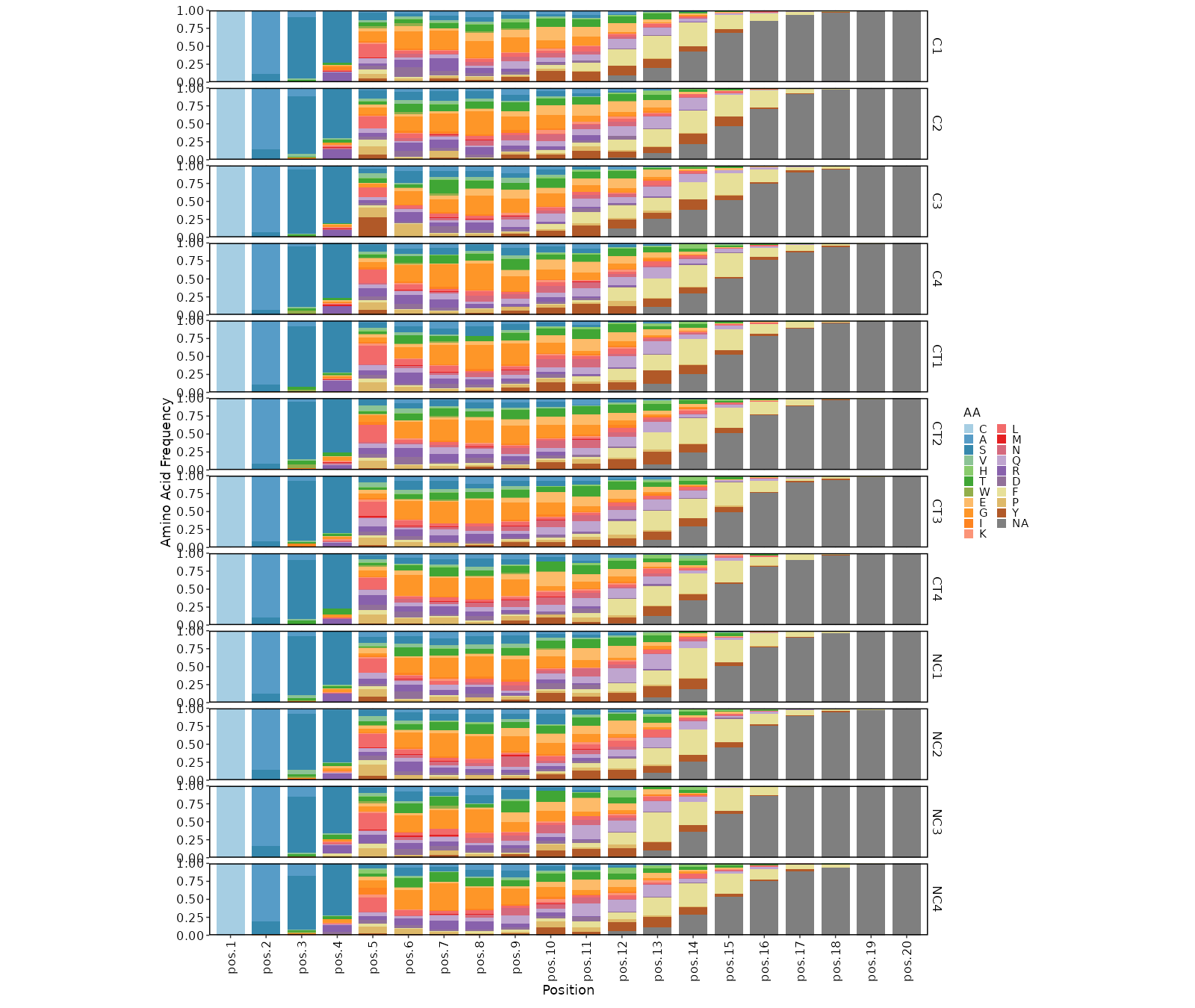 {: width="80%"}
{: width="80%"}
Positional shannon entropy
[ClonalStats.envs.cases."Positional shannon entropy"]
viz_type = "positional"
method = "shannon"
devpars = {width = 1200}
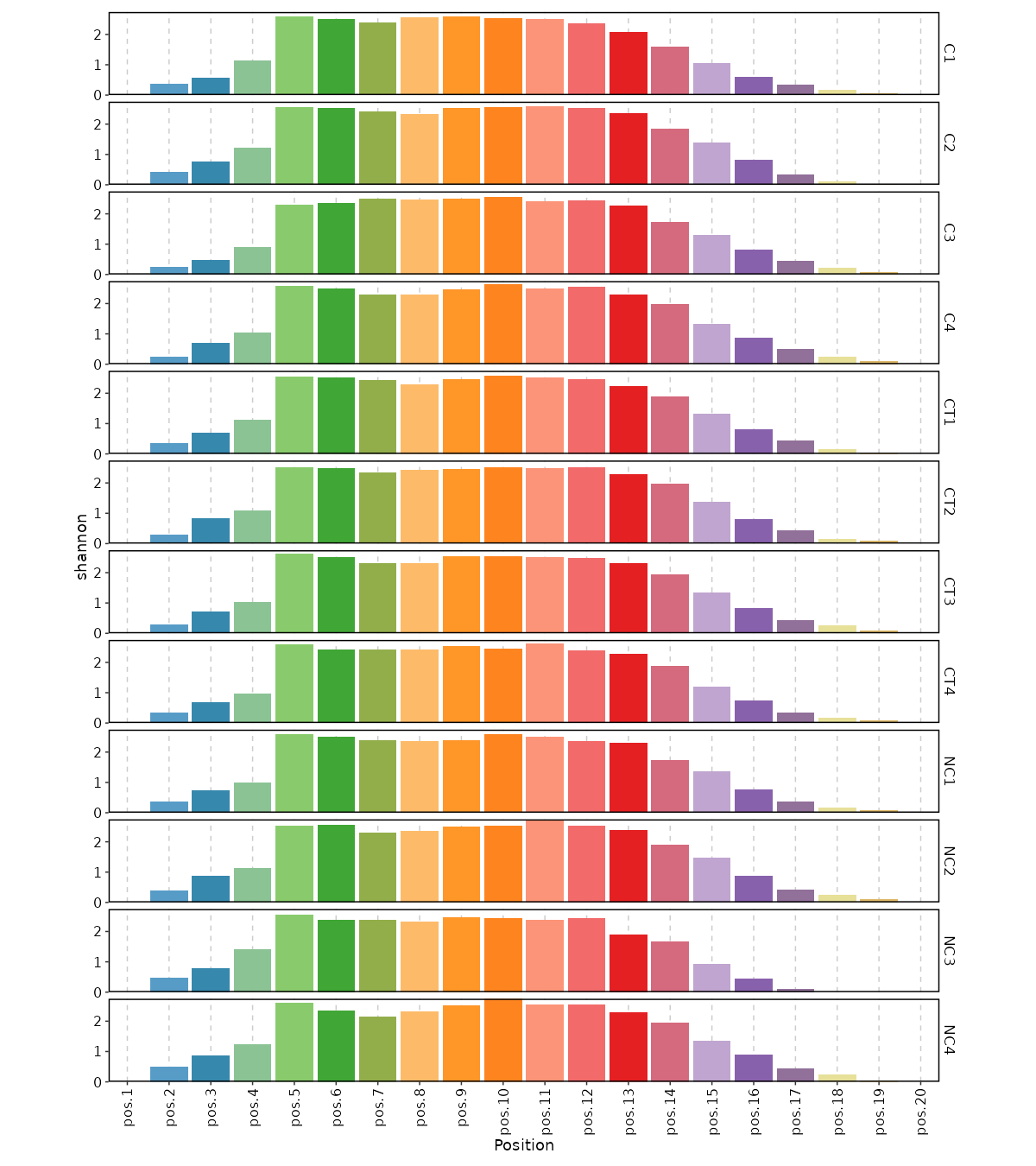 {: width="80%"}
{: width="80%"}
3-Mer Frequency
[ClonalStats.envs.cases."3-Mer Frequency"]
viz_type = "kmer"
k = 3 # default is 3
devpars = {width = 800}
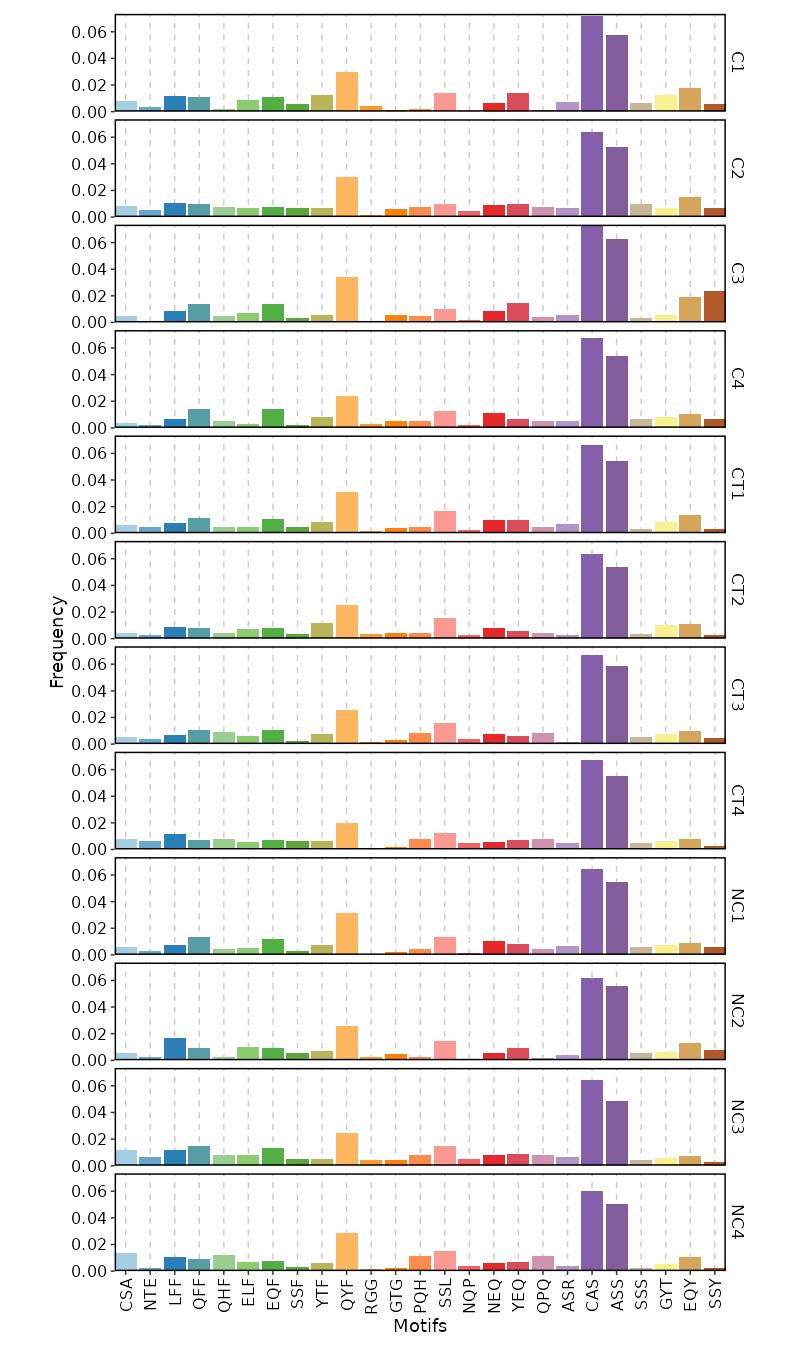 {: width="80%"}
{: width="80%"}
Rarefaction Curve
[ClonalStats.envs.cases."Rarefaction Curve"]
viz_type = "rarefaction"
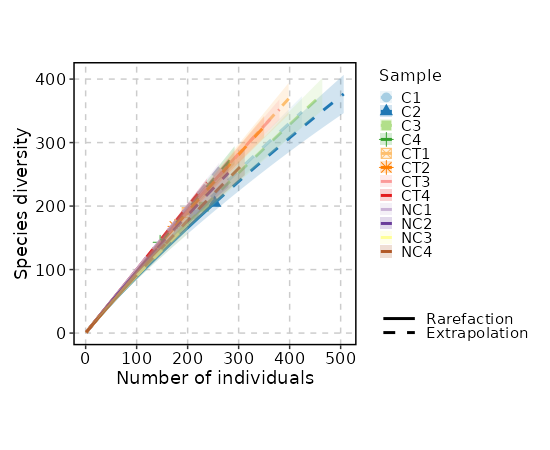 {: width="80%"}
{: width="80%"}
screpfile— ThescRepertoireobject in RDS/qs format
outdir— The output directory containing the plots
cache(type=auto) — Whether to cache the plots.Currently only plots for features are supported, since creating the those plots can be time consuming. IfTrue, the plots will be cached in the job output directory, which will be not cleaned up when job is rerunning.cases(type=json) — The cases to generate the plots if we have multiple cases.The keys are the names of the cases, and the values are the arguments for the plot function. The arguments inenvswill be used if not specified incases, except formutaters. Sections can be specified as the prefix of the case name, separated by::. For example, if you have a case namedClonal Volume::Case1, the plot will be put in the sectionClonal Volume. By default, when there are multiple cases for the same 'viz_type', the name of the 'viz_type' will be used as the default section name (for example, when 'viz_type' is 'volume', the section name will be 'Clonal Volume'). When there is only a single case, the section name will default to 'DEFAULT', which will not be shown in the report.descr— The description of the plot, used to show in the report.devpars(ns) — The parameters for the plotting device.- - width (type=int): The width of the device
- - height (type=int): The height of the device
- - res (type=int): The resolution of the device
more_formats(list) — The extra formats to save the plots in, other than PNG.mutaters(type=json;order=-9) — The mutaters passed todplyr::mutate()to add new variables.When the object loaded formin.screpfileis a list, the mutaters will be applied to each element. The keys are the names of the new variables, and the values are the expressions. When it is aSeuratobject, typically an output ofscRepertoire::combineExpression(), the mutaters will be applied to themeta.data.save_code(flag) — Whether to save the code used to generate the plotsNote that the data directly used to generate the plots will also be saved in anrdafile. Be careful if the data is large as it may take a lot of disk space.save_data(flag) — Whether to save the data used to generate the plot.subset— An expression to subset the data before plotting.Similar tomutaters, it will be applied to each element bydplyr::filter()if the object loaded formin.screpfileis a list; otherwise, it will be applied tosubset(sobj, subset = <expr>)if the object is aSeuratobject.viz_type(choice) — The type of visualization to generate.- - volume: The volume of the clones using
ClonalVolumePlot - - abundance: The abundance of the clones using
ClonalAbundancePlot - - length: The length of the CDR3 sequences using
ClonalLengthPlot - - residency: The residency of the clones using
ClonalResidencyPlot - - stat: The stats of the clones using
ClonalStatPlot - - composition: The composition of the clones using
ClonalCompositionPlot - - overlap: The overlap of the clones using
ClonalOverlapPlot - - diversity: The diversity of the clones using
ClonalDiversityPlot - - geneusage: The gene usage of the clones using
ClonalGeneUsagePlot - - positional: The positional information of the clones using
ClonalPositionalPlot - - kmer: The kmer information of the clones using
ClonalKmerPlot - - rarefaction: The rarefaction curve of the clones using
ClonalRarefactionPlot
- - volume: The volume of the clones using
__init_subclass__()— Do the requirements inferring since we need them to build up theprocess relationship </>from_proc(proc,name,desc,envs,envs_depth,cache,export,error_strategy,num_retries,forks,input_data,order,plugin_opts,requires,scheduler,scheduler_opts,submission_batch)(Type) — Create a subclass of Proc using another Proc subclass or Proc itself</>gc()— GC process for the process to save memory after it's done</>log(level,msg,*args,logger)— Log message for the process</>run()— Init all other properties and jobs</>
pipen.proc.ProcMeta(name, bases, namespace, **kwargs)
Meta class for Proc
__call__(cls,*args,**kwds)(Proc) — Make sure Proc subclasses are singletons</>__instancecheck__(cls,instance)— Override for isinstance(instance, cls).</>__repr__(cls)(str) — Representation for the Proc subclasses</>__subclasscheck__(cls,subclass)— Override for issubclass(subclass, cls).</>register(cls,subclass)— Register a virtual subclass of an ABC.</>
register(cls, subclass)Register a virtual subclass of an ABC.
Returns the subclass, to allow usage as a class decorator.
__instancecheck__(cls, instance)Override for isinstance(instance, cls).
__subclasscheck__(cls, subclass)Override for issubclass(subclass, cls).
__repr__(cls) → strRepresentation for the Proc subclasses
__call__(cls, *args, **kwds)Make sure Proc subclasses are singletons
*args(Any) — and**kwds(Any) — Arguments for the constructor
The Proc instance
from_proc(proc, name=None, desc=None, envs=None, envs_depth=None, cache=None, export=None, error_strategy=None, num_retries=None, forks=None, input_data=None, order=None, plugin_opts=None, requires=None, scheduler=None, scheduler_opts=None, submission_batch=None)
Create a subclass of Proc using another Proc subclass or Proc itself
proc(Type) — The Proc subclassname(str, optional) — The new name of the processdesc(str, optional) — The new description of the processenvs(Mapping, optional) — The arguments of the process, will overwrite parent oneThe items that are specified will be inheritedenvs_depth(int, optional) — How deep to update the envs when subclassed.cache(bool, optional) — Whether we should check the cache for the jobsexport(bool, optional) — When True, the results will be exported to<pipeline.outdir>Defaults to None, meaning only end processes will export. You can set it to True/False to enable or disable exporting for processeserror_strategy(str, optional) — How to deal with the errors- - retry, ignore, halt
- - halt to halt the whole pipeline, no submitting new jobs
- - terminate to just terminate the job itself
num_retries(int, optional) — How many times to retry to jobs once error occursforks(int, optional) — New forks for the new processinput_data(Any, optional) — The input data for the process. Only when this processis a start processorder(int, optional) — The order to execute the new processplugin_opts(Mapping, optional) — The new plugin options, unspecified items will beinherited.requires(Sequence, optional) — The required processes for the new processscheduler(str, optional) — The new shedular to run the new processscheduler_opts(Mapping, optional) — The new scheduler options, unspecified items willbe inherited.submission_batch(int, optional) — How many jobs to be submited simultaneously.
The new process class
__init_subclass__()
Do the requirements inferring since we need them to build up theprocess relationship
run()
Init all other properties and jobs
gc()
GC process for the process to save memory after it's done
log(level, msg, *args, logger=<LoggerAdapter pipen.core (WARNING)>)
Log message for the process
level(int | str) — The log level of the recordmsg(str) — The message to log*args— The arguments to format the messagelogger(LoggerAdapter, optional) — The logging logger