Explore the k-mer frequency of CDR3 sequences.
Arguments
- data
The product of scRepertoire::combineTCR, scRepertoire::combineTCR, or scRepertoire::combineExpression.
- chain
The chain to be analyzed. Default is "TRB".
- clone_call
The column name of the clone call. Default is "aa".
- k
The length of the k-mer. Default is 3.
- top
The number of top k-mers to display. Default is 25.
- group_by
The variable to group the data by. Default is "Sample".
- group_by_sep
The separator to use when combining groupings. Default is "_".
- facet_by
A character vector of column names to facet the plots. Default is NULL.
- split_by
A character vector of column names to split the plots. Default is NULL.
- plot_type
The type of plot to generate. Default is "bar".
"bar": Bar plot.
"line": Line plot.
"heatmap": Heatmap.
- theme_args
A list of arguments to be passed to the ggplot2::theme function.
- aspect.ratio
The aspect ratio of the plot. Default is NULL.
- facet_ncol
The number of columns in the facet grid. Default is NULL.
- ...
Other arguments passed to the specific plot function.
For "bar",
plotthis::BarPlot().For "line",
plotthis::LinePlot().For "heatmap",
plotthis::Heatmap().
Examples
# \donttest{
set.seed(8525)
data(contig_list, package = "scRepertoire")
data <- scRepertoire::combineTCR(contig_list,
samples = c("P17B", "P17L", "P18B", "P18L", "P19B","P19L", "P20B", "P20L"))
data <- scRepertoire::addVariable(data,
variable.name = "Type",
variables = rep(c("B", "L"), 4)
)
data <- scRepertoire::addVariable(data,
variable.name = "Subject",
variables = rep(c("P17", "P18", "P19", "P20"), each = 2)
)
ClonalKmerPlot(data)
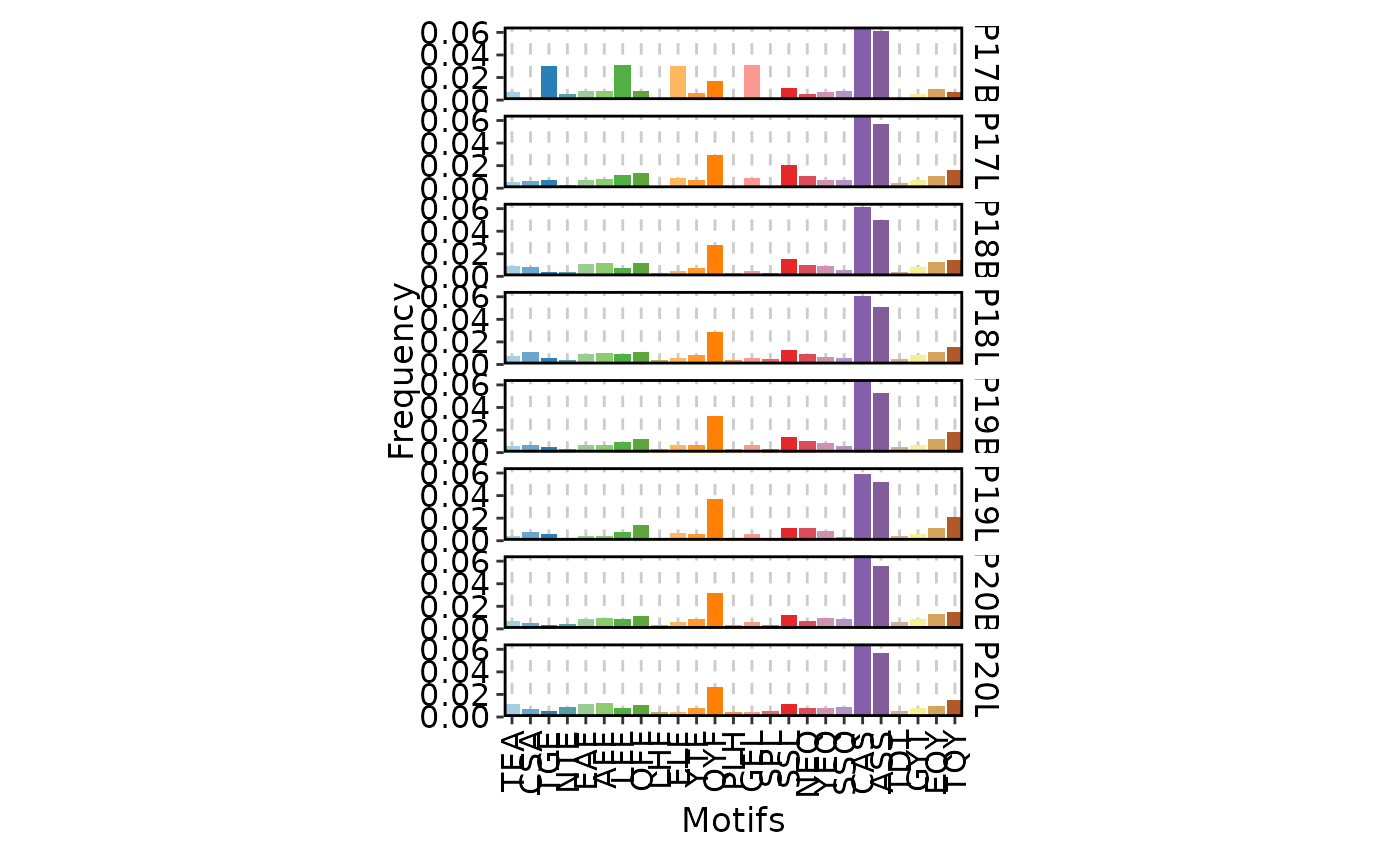 ClonalKmerPlot(data, group_by = "Type")
ClonalKmerPlot(data, group_by = "Type")
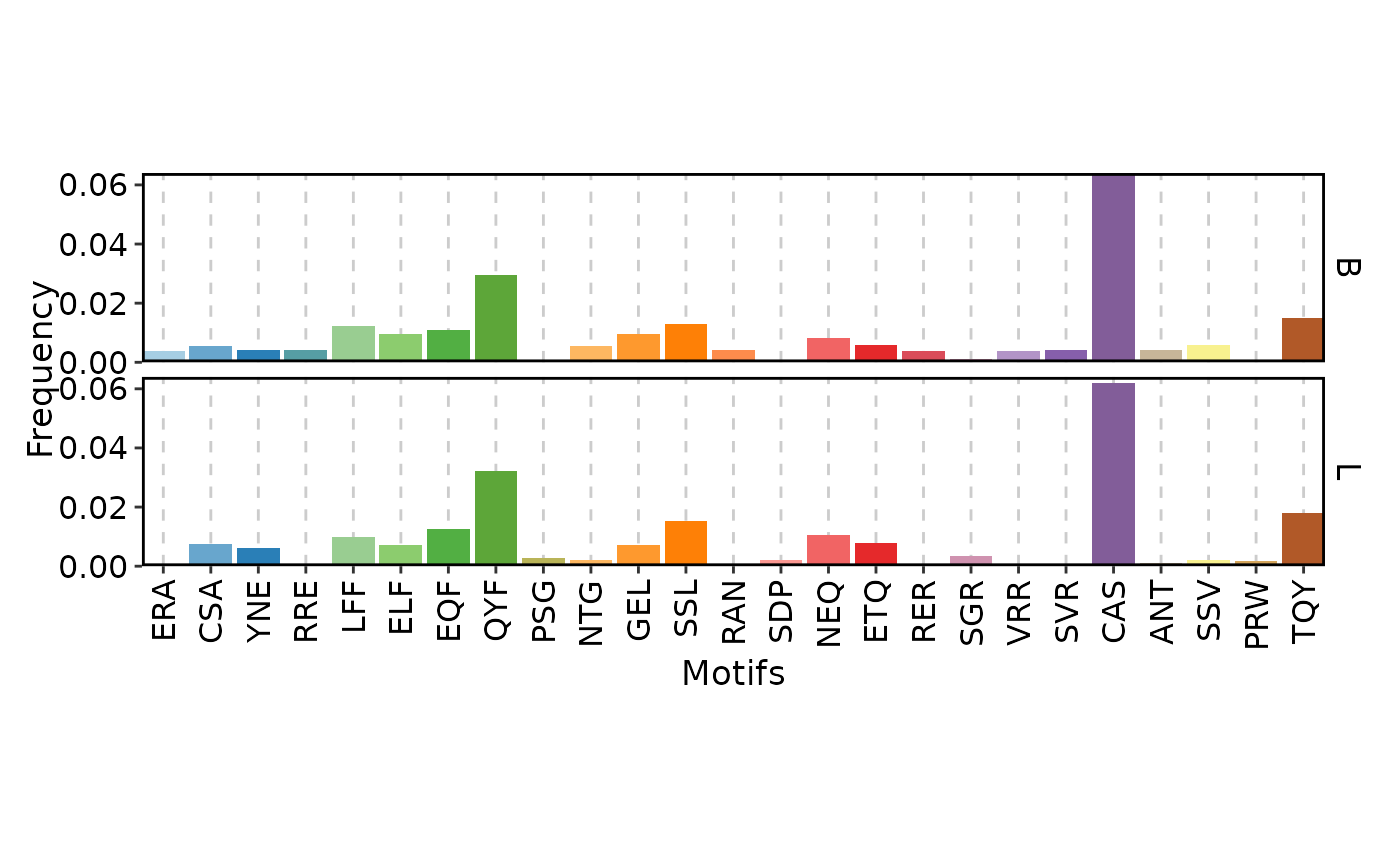 ClonalKmerPlot(data, group_by = "Type", plot_type = "line")
ClonalKmerPlot(data, group_by = "Type", plot_type = "line")
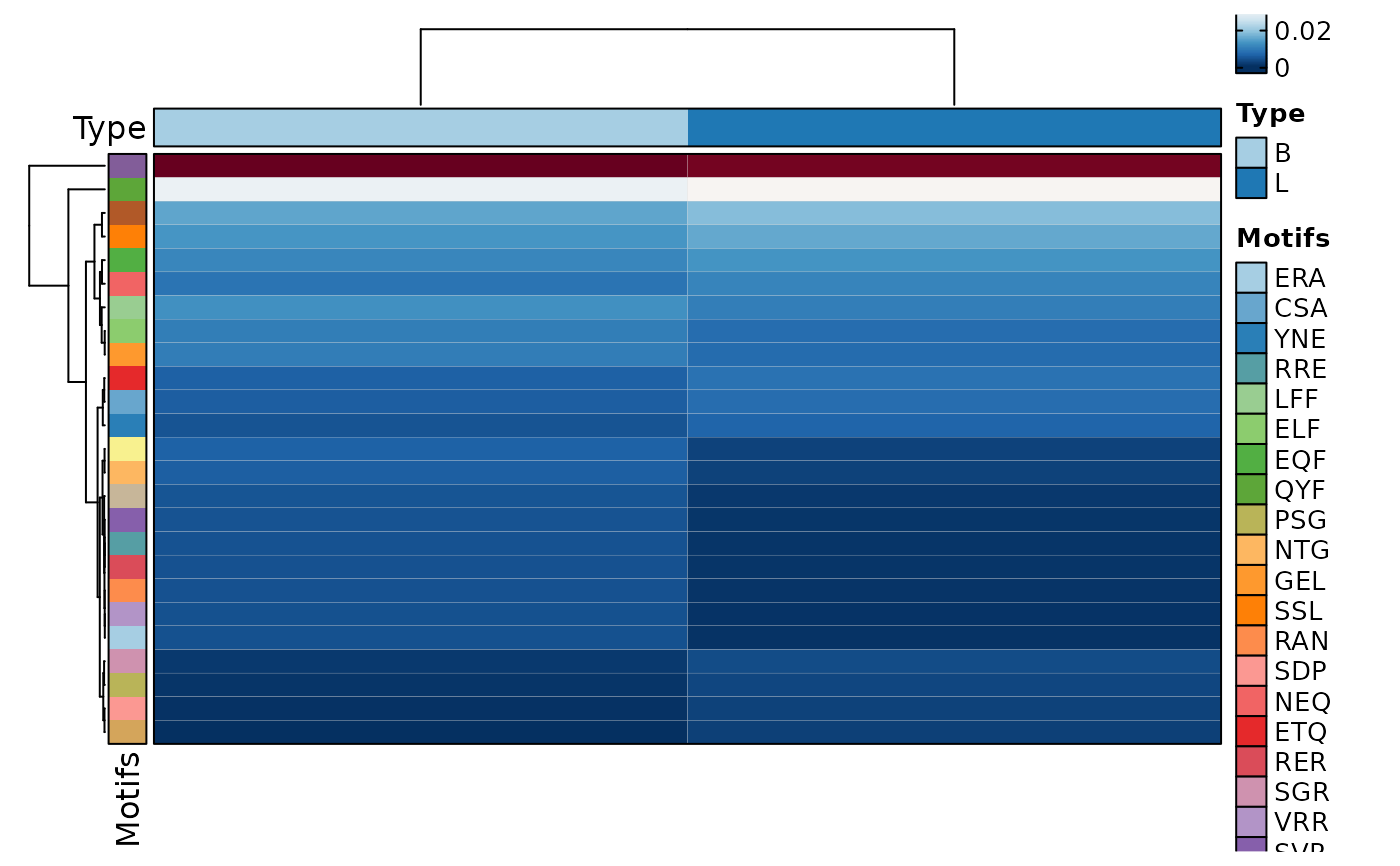
 ClonalKmerPlot(data, group_by = "Type", plot_type = "heatmap")
ClonalKmerPlot(data, group_by = "Type", plot_type = "heatmap")
 # }
# }