Visualizing data with LLMs
Source:vignettes/Visualizing_data_with_LLMs.Rmd
Visualizing_data_with_LLMs.Rmd
library(scplotter)
api_key_set <- !identical(Sys.getenv("OPENAI_API_KEY"), "")Introduction
This vignette demonstrates how to use the scplotter
package to visualize data with AI. The package provides a variety of
functions for visualizing single-cell sequencing data, including
scRNA-seq and scTCR-seq/scBCR-seq data.
Setup LLM provider
scplotter uses tidyprompt to provide a
unified interface for different LLM providers. You can set up your
preferred LLM provider using one of the wrappers
provided by tidyprompt.
# Set up LLM provider
provider <- tidyprompt::llm_provider_openai(
parameters = list(model = "gpt-5-nano", stream = getOption("tidyprompt.stream", TRUE)),
verbose = getOption("tidyprompt.verbose", TRUE),
url = "https://api.openai.com/v1/chat/completions",
api_key = Sys.getenv("OPENAI_API_KEY")
)
chat <- SCPlotterChat$new(provider = provider)Setup the data for visualization
By default, chat will detects the data used for
visualization from the .GlobalEnv and data exported from
the Seurat, SeuratObject, and
scRepertoire packages.
You can also ask to list the available data:
chat$ask("List the available data that can be used for visualization.")
#>
#> Tool identified: ListData
#> Available data objects:
#> - scplotter::cellphonedb_res : A toy example of CellPhoneDB output from LIANA
#> - scplotter::ifnb_sub : A subsetted version of 'ifnb' datasets
#> - scplotter::pancreas_sub : A subsetted version of mouse 'pancreas' datasets
#> - Seurat::cc.genes : Cell cycle genes
#> - Seurat::cc.genes.updated.2019 : Cell cycle genes: 2019 update
#> - SeuratObject::pbmc_small : A small example version of the PBMC dataset
#> - scRepertoire::contig_list : A list of 8 single-cell T cell receptor sequences runs.
#> - scRepertoire::mini_contig_list : Processed subset of 'contig_list'
#> - scRepertoire::scRep_example : A Seurat object of 500 single T cells,
# or you can do it explicitly
# chat$list_data()To set up the data manually, you can use the set_data()
method.
chat$set_data(scplotter::cellphonedb_res)
# To let the LLM to detect the data from the prompt again:
chat$set_data(NULL)To use your own data, you can either set the data manually or use the
set_data() method or you can load the data in the global
environment and mention it in your prompt.
List the available tools
You can list the available functions by using the
list_tools() method.
chat$list_tools()
#> Available tools:
#> - gt : Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#> - CellDimPlot : Cell Dimension Reduction Plot
#> This function creates a dimension reduction plot for a Seurat object
#> a Giotto object, a path to an .h5ad file or an opened H5File by hdf5r package.
#> It allows for various customizations such as grouping by metadata,
#> adding edges between cell neighbors, highlighting specific cells, and more.
#> This function is a wrapper around plotthis::DimPlot() , which provides a
#> flexible way to visualize cell clusters in reduced dimensions. This function
#> extracts the necessary data from the Seurat or Giotto object and passes it to
#> plotthis::DimPlot() .
#> - ClonalVolumePlot : ClonalVolumePlot
#> ClonalVolumePlot
#> - ClonalRarefactionPlot : ClonalRarefactionPlot
#> Plot the rarefaction curves
#> - ClonalStatPlot : ClonalStatPlot
#> Visualize the statistics of the clones.
#> - ClonalCompositionPlot : ClonalCompositionPlot
#> Plot the composition of the clones in different samples/groups.
#> - EnrichmentPlot : Enrichment Plot
#> This function generates various types of plots for enrichment (over-representation) analysis.
#> - eq : Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#> - CellVelocityPlot : Cell Velocity Plot
#> This function creates a cell velocity plot for a Seurat object,
#> a Giotto object, a path to an .h5ad file or an opened H5File by hdf5r package.
#> It allows for various customizations such as grouping by metadata,
#> adding edges between cell neighbors, highlighting specific cells, and more.
#> This function is a wrapper around plotthis::VelocityPlot() , which provides a
#> flexible way to visualize cell velocities in reduced dimensions. This function
#> extracts the cell embeddings and velocity embeddings from the Seurat or Giotto object
#> and passes them to plotthis::VelocityPlot() .
#> - SpatFeaturePlot : Plot features for spatial data
#> The features can include expression, dimension reduction components, metadata, etc
#> - shared : Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#> - CellStatPlot : Cell statistics plot
#> This function creates a plot to visualize the statistics of cells in a Seurat object, a Giotto object,
#> a path to an .h5ad file or an opened H5File by hdf5r package.
#> It can create various types of plots, including bar plots, circos plots, pie charts, pies (heatmap with cell_type = 'pie'), ring/donut plots, trend plots
#> area plots, sankey/alluvial plots, heatmaps, radar plots, spider plots, violin plots, and box plots.
#> The function allows for grouping, splitting, and faceting the data based on metadata columns.
#> It also supports calculating fractions of cells based on specified groupings.#'
#> - ne : Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#> - SpatDimPlot : Plot categories for spatial data
#> Plot categories for spatial data
#> - GSEASummaryPlot : Objects exported from other packages
#> These objects are imported from other packages. Follow the links
#> below to see their documentation.
#>
#>
#> plotthis GSEAPlot , GSEASummaryPlot
#> - CCCPlot : Cell-Cell Communication Plot
#> Plot the cell-cell communication.
#> See also:
#>
#> The review: https://www.sciencedirect.com/science/article/pii/S2452310021000081
#> The LIANA package: https://liana-py.readthedocs.io/en/latest/notebooks/basic_usage.html#Tileplot
#> The CCPlotR package: https://github.com/Sarah145/CCPlotR
#>
#> - ClonalResidencyPlot : ClonalResidencyPlot
#> Plot the residency of the clones in different samples.
#> - ClustreePlot : Clustree plot
#> This function generates a clustree plot from a data frame or a Seurat object.
#> - ClonalKmerPlot : ClonalKmerPlot
#> Explore the k-mer frequency of CDR3 sequences.
#> - ClonalPositionalPlot : ClonalPositionalPlot
#> Visualize the positional entropy, property or amino acid frequency of CDR3 sequences.
#> - uniq : Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#> - top : Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#> - le : Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#> - MarkersPlot : Visualize Markers
#> Plot markers, typically identified by Seurat::FindMarkers() or Seurat::FindAllMarkers() .
#> - ClonalOverlapPlot : ClonalOverlapPlot
#> Plot the overlap of the clones in different samples/groups.
#> - ge : Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#> - ClonalDynamicsPlot : ClonalDynamicsPlot
#> This function is deprecated. Please use ClonalStatPlot() instead.
#> - ClonalLengthPlot : ClonalLengthPlot
#> Plot the length distribution of the CDR3 sequences
#> - and : Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#> - ClonalAbundancePlot : ClonalAbundancePlot
#> Plot the count or density of the clones at different abundance levels.
#> - sel : Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#> - ClonalDiversityPlot : ClonalDiversityPlot
#> Plot the clonal diversities of the samples/groups.
#> - FeatureStatPlot : Feature statistic plot
#> This function creates various types of feature statistic plots for a Seurat object, a Giotto object,
#> a path to an .h5ad file or an opened H5File by hdf5r package.
#> It allows for plotting features such as gene expression, scores, or other metadata across different groups or conditions.
#> The function supports multiple plot types including violin, box, bar, ridge, dimension reduction, correlation, heatmap, and dot plots.
#> It can also handle multiple features and supports faceting, splitting, and grouping by metadata columns.
#> - ClonalGeneUsagePlot : ClonalGeneUsagePlot
#> ClonalGeneUsagePlot
#> - lt : Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#> - GSEAPlot : Objects exported from other packages
#> These objects are imported from other packages. Follow the links
#> below to see their documentation.
#>
#>
#> plotthis GSEAPlot , GSEASummaryPlot
#> - or : Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#> - ListTools : List all available tools
#> List all available tools that can be used to handle the chat request.
#> - ListData : List all available data objects
#> List all available data objects that can be used to handle the chat request.
# or you can ask the LLM to list the available functions
# chat$ask("List the available functions for visualizing data.")The tool used for the visualization is determined by the LLM automatically from your prompt.
Visualize the data
You can visualize the data by using the ask() method.
The LLM will automatically detect the data and the function to be used
for visualization.
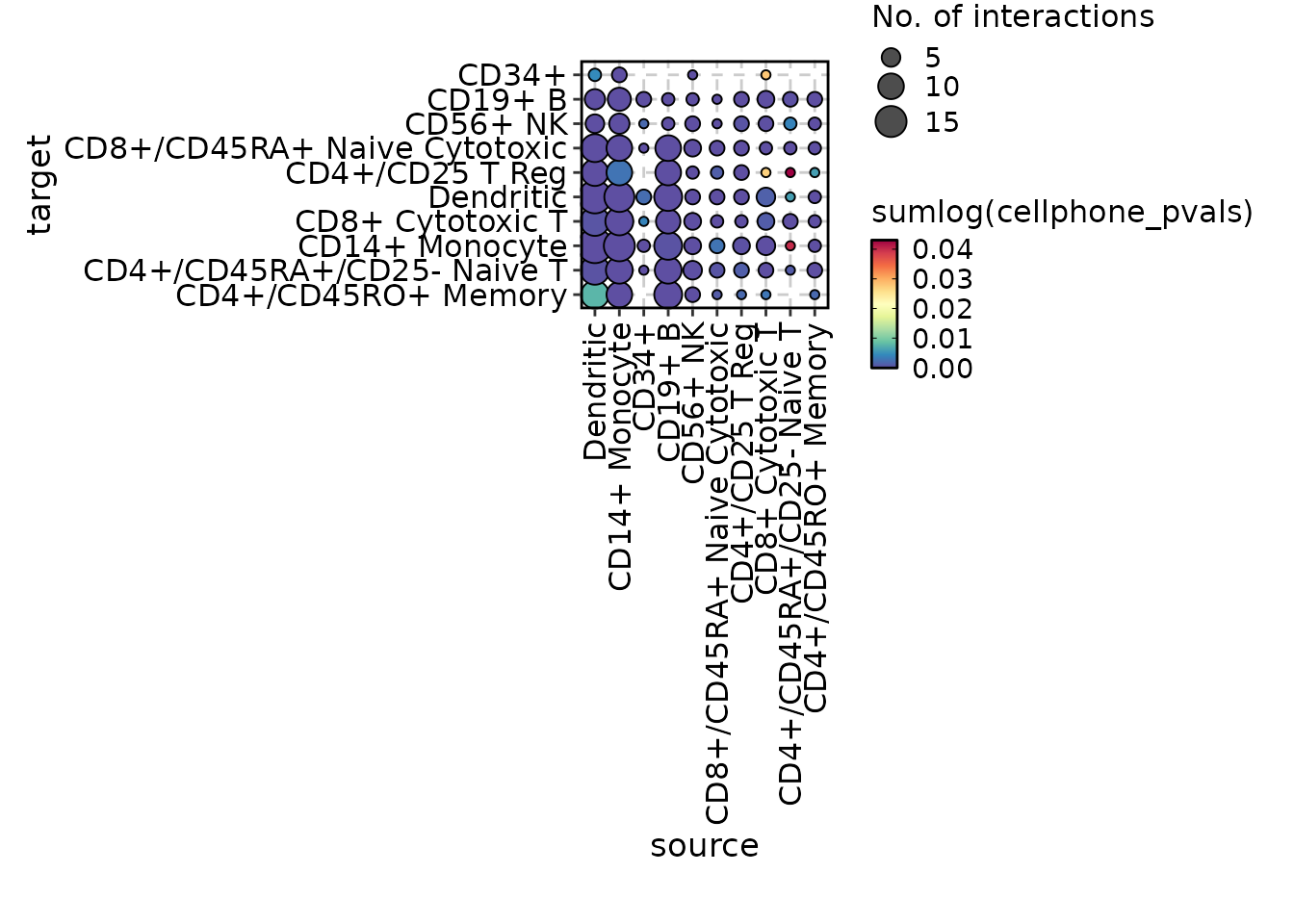
chat$ask("Generate a cell-cell communication plot for the cellphonedb_res data.")
#>
#> Tool identified: CCCPlot
#>
#> Data object identified: scplotter::cellphonedb_res
#> Warning in wrap$modify_fn(prompt_text, llm_provider): The 'skimr' package is
#> required to skim dataframes. Skim summary of dataframes currently not shown in
#> prompt
#> Code ran:
#> CCCPlot(data = cellphonedb_res)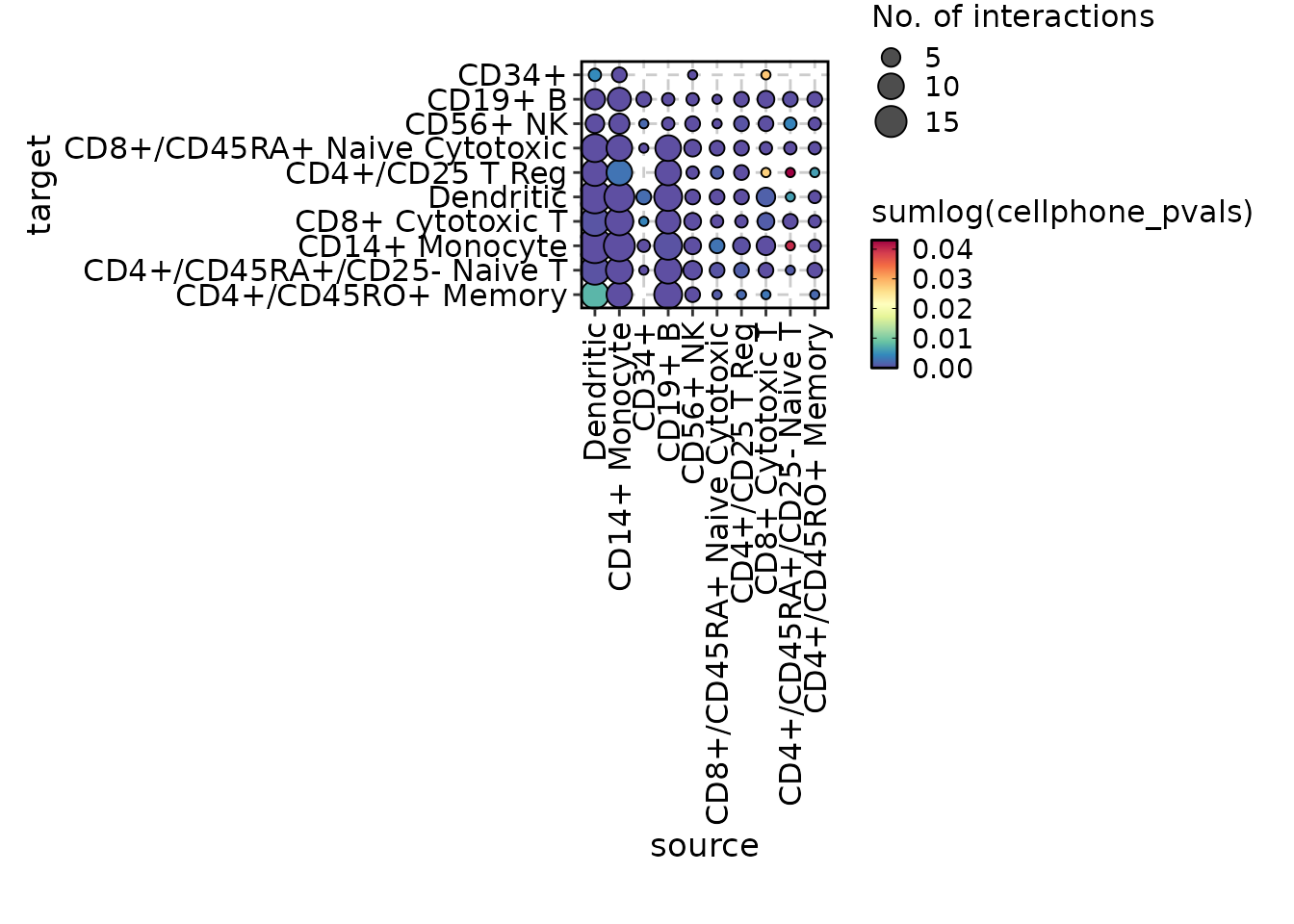
# Previous conversation is memorized
chat$ask("Do a heatmap instead")
#>
#> Tool identified: CCCPlot
#>
#> Data object identified: scplotter::cellphonedb_res
#> Warning in wrap$modify_fn(prompt_text, llm_provider): The 'skimr' package is
#> required to skim dataframes. Skim summary of dataframes currently not shown in
#> prompt
#> Code ran:
#> CCCPlot(cellphonedb_res, plot_type = "heatmap")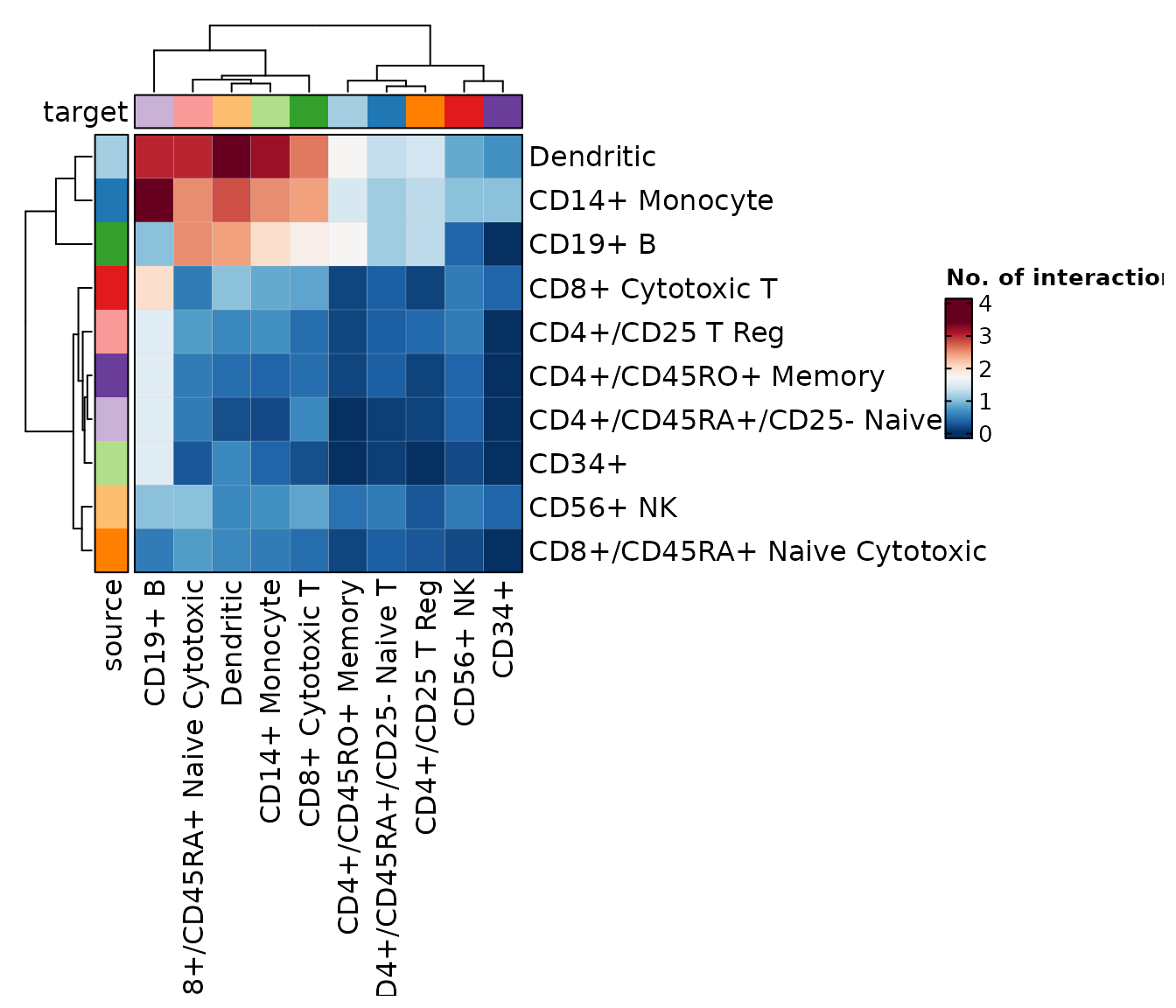
chat$ask("Add a proper title to the plot")
#>
#> Tool identified: CCCPlot
#>
#> Data object identified: scplotter::cellphonedb_res
#> Warning in wrap$modify_fn(prompt_text, llm_provider): The 'skimr' package is
#> required to skim dataframes. Skim summary of dataframes currently not shown in
#> prompt
#> Code ran:
#> CCCPlot(data = cellphonedb_res, plot_type = "heatmap", title = "Cell-Cell Communication Heatmap")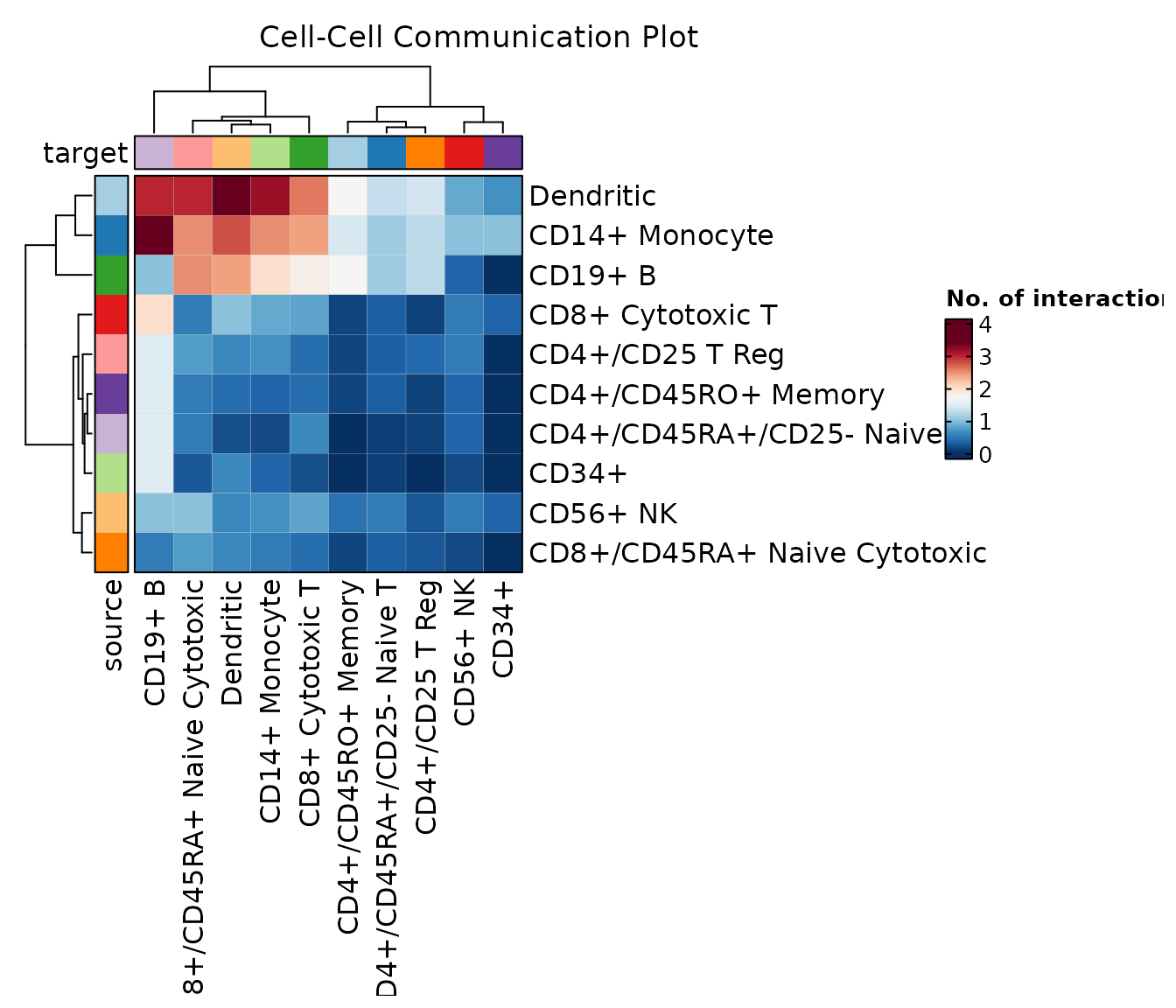
# To fetch the previous conversation
# Note that the response from the LLM is simplified in the history
chat$get_history()
#> [1] "User: Generate a cell-cell communication plot for the cellphonedb_res data."
#> [2] "Assistant: tool - CCCPlot; data - scplotter::cellphonedb_res; code - CCCPlot(data = cellphonedb_res)"
#> [3] "User: Do a heatmap instead"
#> [4] "Assistant: tool - CCCPlot; data - scplotter::cellphonedb_res; code - CCCPlot(cellphonedb_res, plot_type = \"heatmap\")"
#> [5] "User: Add a proper title to the plot"
#> [6] "Assistant: tool - CCCPlot; data - scplotter::cellphonedb_res; code - CCCPlot(data = cellphonedb_res, plot_type = \"heatmap\", title = \"Cell-Cell Communication Heatmap\")"
# To clear the history
chat$clear_history()Debug and improve the prompt
You can set verbose to TRUE for all
conversations when constructing the chat object. This will
print the prompt and the response from the LLM.
chat <- SCPlotterChat$new(
provider = provider,
verbose = TRUE
)
chat$ask("Generate a cell-cell communication plot for the cellphonedb_res data.")
#> --- Sending request to LLM provider (gpt-5-nano): ---
#> Objective: Select the most appropriate tool to handle the user's request while preserving conversational context. If the user refines or changes how the previous result should be visualized (e.g., asks for a different plot type), continue with the last plotting tool used unless they explicitly name a different tool.
#>
#> Decision Process:
#> - If the user explicitly names a tool, output that tool.
#> - Else, if the request appears to refine the previous output (e.g., "do X instead", "make it a heatmap/dot/bar/etc", "change to ...", "add ... to", "same plot but ..."), select the last tool mentioned in the chat history.
#> - Else, analyze the current user request; if there is a clear, unambiguous match to a single tool, select that tool.
#> - Else, use the last mentioned tool from the chat history.
#> - If no tool is found, respond with "None".
#>
#> Response Format: Provide only the name of the selected tool, or "None" if no tool applies.
#>
#> User Request:
#> Generate a cell-cell communication plot for the cellphonedb_res data.
#>
#> Available Tools:
#> - gt: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - CellDimPlot: Cell Dimension Reduction Plot
#> This function creates a dimension reduction plot for a Seurat object
#> a Giotto object, a path to an .h5ad file or an opened H5File by hdf5r package.
#> It allows for various customizations such as grouping by metadata,
#> adding edges between cell neighbors, highlighting specific cells, and more.
#> This function is a wrapper around plotthis::DimPlot() , which provides a
#> flexible way to visualize cell clusters in reduced dimensions. This function
#> extracts the necessary data from the Seurat or Giotto object and passes it to
#> plotthis::DimPlot() .
#>
#> - ClonalVolumePlot: ClonalVolumePlot
#> ClonalVolumePlot
#>
#> - ClonalRarefactionPlot: ClonalRarefactionPlot
#> Plot the rarefaction curves
#>
#> - ClonalStatPlot: ClonalStatPlot
#> Visualize the statistics of the clones.
#>
#> - ClonalCompositionPlot: ClonalCompositionPlot
#> Plot the composition of the clones in different samples/groups.
#>
#> - EnrichmentPlot: Enrichment Plot
#> This function generates various types of plots for enrichment (over-representation) analysis.
#>
#> - eq: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - CellVelocityPlot: Cell Velocity Plot
#> This function creates a cell velocity plot for a Seurat object,
#> a Giotto object, a path to an .h5ad file or an opened H5File by hdf5r package.
#> It allows for various customizations such as grouping by metadata,
#> adding edges between cell neighbors, highlighting specific cells, and more.
#> This function is a wrapper around plotthis::VelocityPlot() , which provides a
#> flexible way to visualize cell velocities in reduced dimensions. This function
#> extracts the cell embeddings and velocity embeddings from the Seurat or Giotto object
#> and passes them to plotthis::VelocityPlot() .
#>
#> - SpatFeaturePlot: Plot features for spatial data
#> The features can include expression, dimension reduction components, metadata, etc
#>
#> - shared: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - CellStatPlot: Cell statistics plot
#> This function creates a plot to visualize the statistics of cells in a Seurat object, a Giotto object,
#> a path to an .h5ad file or an opened H5File by hdf5r package.
#> It can create various types of plots, including bar plots, circos plots, pie charts, pies (heatmap with cell_type = 'pie'), ring/donut plots, trend plots
#> area plots, sankey/alluvial plots, heatmaps, radar plots, spider plots, violin plots, and box plots.
#> The function allows for grouping, splitting, and faceting the data based on metadata columns.
#> It also supports calculating fractions of cells based on specified groupings.#'
#>
#> - ne: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - SpatDimPlot: Plot categories for spatial data
#> Plot categories for spatial data
#>
#> - GSEASummaryPlot: Objects exported from other packages
#> These objects are imported from other packages. Follow the links
#> below to see their documentation.
#>
#>
#> plotthis GSEAPlot , GSEASummaryPlot
#>
#> - CCCPlot: Cell-Cell Communication Plot
#> Plot the cell-cell communication.
#> See also:
#>
#> The review: https://www.sciencedirect.com/science/article/pii/S2452310021000081
#> The LIANA package: https://liana-py.readthedocs.io/en/latest/notebooks/basic_usage.html#Tileplot
#> The CCPlotR package: https://github.com/Sarah145/CCPlotR
#>
#>
#> - ClonalResidencyPlot: ClonalResidencyPlot
#> Plot the residency of the clones in different samples.
#>
#> - ClustreePlot: Clustree plot
#> This function generates a clustree plot from a data frame or a Seurat object.
#>
#> - ClonalKmerPlot: ClonalKmerPlot
#> Explore the k-mer frequency of CDR3 sequences.
#>
#> - ClonalPositionalPlot: ClonalPositionalPlot
#> Visualize the positional entropy, property or amino acid frequency of CDR3 sequences.
#>
#> - uniq: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - top: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - le: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - MarkersPlot: Visualize Markers
#> Plot markers, typically identified by Seurat::FindMarkers() or Seurat::FindAllMarkers() .
#>
#> - ClonalOverlapPlot: ClonalOverlapPlot
#> Plot the overlap of the clones in different samples/groups.
#>
#> - ge: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - ClonalDynamicsPlot: ClonalDynamicsPlot
#> This function is deprecated. Please use ClonalStatPlot() instead.
#>
#> - ClonalLengthPlot: ClonalLengthPlot
#> Plot the length distribution of the CDR3 sequences
#>
#> - and: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - ClonalAbundancePlot: ClonalAbundancePlot
#> Plot the count or density of the clones at different abundance levels.
#>
#> - sel: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - ClonalDiversityPlot: ClonalDiversityPlot
#> Plot the clonal diversities of the samples/groups.
#>
#> - FeatureStatPlot: Feature statistic plot
#> This function creates various types of feature statistic plots for a Seurat object, a Giotto object,
#> a path to an .h5ad file or an opened H5File by hdf5r package.
#> It allows for plotting features such as gene expression, scores, or other metadata across different groups or conditions.
#> The function supports multiple plot types including violin, box, bar, ridge, dimension reduction, correlation, heatmap, and dot plots.
#> It can also handle multiple features and supports faceting, splitting, and grouping by metadata columns.
#>
#> - ClonalGeneUsagePlot: ClonalGeneUsagePlot
#> ClonalGeneUsagePlot
#>
#> - lt: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - GSEAPlot: Objects exported from other packages
#> These objects are imported from other packages. Follow the links
#> below to see their documentation.
#>
#>
#> plotthis GSEAPlot , GSEASummaryPlot
#>
#> - or: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - ListTools: List all available tools
#> List all available tools that can be used to handle the chat request.
#>
#> - ListData: List all available data objects
#> List all available data objects that can be used to handle the chat request.
#> --- Receiving response from LLM provider: ---
#> CCCPlot
#> Tool identified: CCCPlot
#> --- Sending request to LLM provider (gpt-5-nano): ---
#> Objective: Identify the data object to be used.
#>
#> Decision Process:
#> - If the user explicitly names a data object, output that data object name. If the name does not match exactly the available ones, use the one that is mostly related.
#> - Else, if the request appears to refine the previous output (e.g., "do X instead", "make it a heatmap/dot/bar/etc", "change to ...", "add ... to", "same plot but ..."), select the last dataset name mentioned in the chat history.
#> - Else, analyze the current user request; if there is a clear, unambiguous match to a dataset, select that data object.
#> - Else, use the last mentioned data object from the chat history.
#> - If no data object is found, respond with "None".
#>
#> Response Format: Provide only the name of the selected data object, or "None" if no tool applies. The response should be unquoted.
#>
#> User Request:
#> Generate a cell-cell communication plot for the cellphonedb_res data.
#>
#> Available Data Objects:
#> - scplotter::cellphonedb_res: A toy example of CellPhoneDB output from LIANA
#> - scplotter::ifnb_sub: A subsetted version of 'ifnb' datasets
#> - scplotter::pancreas_sub: A subsetted version of mouse 'pancreas' datasets
#> - Seurat::cc.genes: Cell cycle genes
#> - Seurat::cc.genes.updated.2019: Cell cycle genes: 2019 update
#> - SeuratObject::pbmc_small: A small example version of the PBMC dataset
#> - scRepertoire::contig_list: A list of 8 single-cell T cell receptor sequences runs.
#> - scRepertoire::mini_contig_list: Processed subset of 'contig_list'
#> - scRepertoire::scRep_example: A Seurat object of 500 single T cells,
#> --- Receiving response from LLM provider: ---
#> scplotter::cellphonedb_res
#> Data object identified: scplotter::cellphonedb_res
#> Warning in wrap$modify_fn(prompt_text, llm_provider): The 'skimr' package is
#> required to skim dataframes. Skim summary of dataframes currently not shown in
#> prompt
#> --- Sending request to LLM provider (gpt-5-nano): ---#> Objective: Generate the R code to run the specified tool using the specified data object based on the user's request and the provided tool information.
#>
#> Decision Process:
#> - Analyze the user's request to understand what needs to be done with the specified tool and data object.
#> - When use the data object name, do not quote it.
#> - Refer to the provided tool information to understand the tool's usage, arguments, and examples.
#> - Consider any specific parameters or options mentioned in the user's request that need to be included in the tool call.
#> - If the user's request is ambiguous or lacks necessary details, refer to the chat history for additional context that may help clarify the intended use of the tool and data object.
#> - Construct the R code that correctly calls the tool with the specified data object, ensuring that all necessary arguments are included and correctly formatted.
#>
#> Response Format: Provide only the valid R code wrapped between "```r" and "```" to run the tool.
#>
#> Tool to be used: CCCPlot
#> Data object to be used: cellphonedb_res
#>
#> User Request:
#> Generate a cell-cell communication plot for the cellphonedb_res data.
#>
#> Tool Information:
#> - title
#> Cell-Cell Communication Plot
#> - description
#>
#> Plot the cell-cell communication.
#> See also:
#>
#> The review: https://www.sciencedirect.com/science/article/pii/S2452310021000081
#> The LIANA package: https://liana-py.readthedocs.io/en/latest/notebooks/basic_usage.html#Tileplot
#> The CCPlotR package: https://github.com/Sarah145/CCPlotR
#>
#>
#> - usage
#>
#> CCCPlot(
#> data,
#> plot_type = c("dot", "network", "chord", "circos", "heatmap", "sankey", "alluvial",
#> "box", "violin", "ridge"),
#> method = c("aggregation", "interaction"),
#> magnitude = waiver(),
#> specificity = waiver(),
#> magnitude_agg = length,
#> magnitude_name = "No. of interactions",
#> meta_specificity = "sumlog",
#> split_by = NULL,
#> x_text_angle = 90,
#> link_curvature = 0.2,
#> link_alpha = 0.6,
#> facet_by = NULL,
#> show_row_names = TRUE,
#> show_column_names = TRUE,
#> ...
#> )
#>
#> - arguments
#> - data: A data frame with the cell-cell communication data.
#> A typical data frame should have the following columns:
#>
#> source The source cell type.
#> target The target cell type.
#> ligand The ligand gene.
#> receptor The receptor gene.
#> ligand_means The mean expression of the ligand gene per cell type.
#> receptor_means The mean expression of the receptor gene per cell type.
#> ligand_props The proportion of cells that express the entity.
#> receptor_props The proportion of cells that express the entity.
#> <magnitude> The magnitude of the communication.
#> <specificity> The specificity of the communication.
#> Depends on the plot_type, some columns are optional. But the source, target,
#> ligand, receptor and <magnitude> are required.
#>
#> - plot_type: The type of plot to use. Default is "dot".
#> Possible values are "network", "chord", "circos", "heatmap", "sankey", "alluvial", "dot",
#> "box", "violin" and "ridge".
#> For "box", "violin" and "ridge", the method should be "interaction".
#>
#> network: A network plot with the source and target cells as the nodes and the communication as the edges.
#> chord: A chord plot with the source and target cells as the nodes and the communication as the chords.
#> circos: Alias of "chord".
#> heatmap: A heatmap plot with the source and target cells as the rows and columns.
#> sankey: A sankey plot with the source and target cells as the nodes and the communication as the flows.
#> alluvial: Alias of "sankey".
#> dot: A dot plot with the source and target cells as the nodes and the communication as the dots.
#> box: Box plots for source cell types. Each x is a target cell type and the values will be
#> the interaction strengths of the ligand-receptor pairs.
#> violin: Violin plots for source cell types. Each x is a target cell type and the values will be
#> the interaction strengths of the ligand-receptor pairs.
#> ridge: Ridge plots for source cell types. Each row is a target cell type and the values will be
#> the interaction strengths of the ligand-receptor pairs.
#>
#> - method: The method to determine the plot entities.
#>
#> aggregation: Aggregate the ligand-receptor pairs interactions for each source-target pair.
#> Only the source / target pairs will be plotted.
#> interaction: Plot the ligand-receptor pairs interactions directly.
#> The ligand-receptor pairs will also be plotted.
#>
#> - magnitude: The column name in the data to use as the magnitude of the communication.
#> By default, the second last column will be used.
#> See li.mt.show_methods() for the available methods in LIANA.
#> or https://liana-py.readthedocs.io/en/latest/notebooks/basic_usage.html#Tileplot
#> - specificity: The column name in the data to use as the specificity of the communication.
#> By default, the last column will be used.
#> If the method doesn't have a specificity, set it to NULL.
#> - magnitude_agg: A function to aggregate the magnitude of the communication.
#> Default is length.
#> - magnitude_name: The name of the magnitude in the plot.
#> Default is "No. of interactions".
#> - meta_specificity: The method to calculate the specificity when there are multiple
#> ligand-receptor pairs interactions. Default is "sumlog".
#> It should be one of the methods in the metap package.
#> Current available methods are:
#>
#> invchisq: Combine p values using the inverse chi squared method
#> invt: Combine p values using the inverse t method
#> logitp: Combine p values using the logit method
#> meanp: Combine p values by the mean p method
#> meanz: Combine p values using the mean z method
#> sumlog: Combine p-values by the sum of logs (Fisher's) method
#> sump: Combine p-values using the sum of p (Edgington's) method
#> two2one: Convert two-sided p-values to one-sided
#> votep: Combine p-values by the vote counting method
#> wilkinsonp: Combine p-values using Wilkinson's method
#>
#> - split_by: A character vector of column names to split the plots. Default is NULL.
#> - x_text_angle: The angle of the x-axis text. Default is 90.
#> Only used when plot_type is "dot".
#> - link_curvature: The curvature of the links. Default is 0.2.
#> Only used when plot_type is "network".
#> - link_alpha: The transparency of the links. Default is 0.6.
#> Only used when plot_type is "network".
#> - facet_by: A character vector of column names to facet the plots. Default is NULL.
#> It should always be NULL.
#> - show_row_names: Whether to show the row names in the heatmap. Default is TRUE.
#> Only used when plot_type is "heatmap".
#> - show_column_names: Whether to show the column names in the heatmap. Default is TRUE.
#> Only used when plot_type is "heatmap".
#> - ...: Other arguments passed to the specific plot function.
#>
#> For Network, see plotthis::Network().
#>
#> [...] can be:
#> - links: A data frame containing the links between nodes.
#> - nodes: A data frame containing the nodes.
#> This is optional. The names of the nodes are extracted from the links data frame.
#> If "@nodes" is provided, the nodes data frame will be extracted from the attribute nodes of the links data frame.
#> - split_by_sep: The separator for multiple split_by columns. See split_by
#> - split_nodes: A logical value specifying whether to split the nodes data.
#> If TRUE, the nodes data will also be split by the split_by column.
#> - from: A character string specifying the column name of the links data frame for the source nodes.
#> Default is the first column of the links data frame.
#> - from_sep: A character string to concatenate the columns in from, if multiple columns are provided.
#> - to: A character string specifying the column name of the links data frame for the target nodes.
#> Default is the second column of the links data frame.
#> - to_sep: A character string to concatenate the columns in to, if multiple columns are provided.
#> - node_by: A character string specifying the column name of the nodes data frame for the node names.
#> Default is the first column of the nodes data frame.
#> - node_by_sep: A character string to concatenate the columns in node_by, if multiple columns are provided.
#> - link_weight_by: A numeric value or a character string specifying the column name of the links data frame for the link weight.
#> If a numeric value is provided, all links will have the same weight.
#> This determines the width of the links.
#> - link_weight_name: A character string specifying the name of the link weight in the legend.
#> - link_type_by: A character string specifying the type of the links.
#> This can be "solid", "dashed", "dotted", or a column name from the links data frame.
#> It has higher priority when it is a column name.
#> - link_type_name: A character string specifying the name of the link type in the legend.
#> - node_size_by: A numeric value or a character string specifying the column name of the nodes data frame for the node size.
#> If a numeric value is provided, all nodes will have the same size.
#> - node_size_name: A character string specifying the name of the node size in the legend.
#> - node_color_by: A character string specifying the color of the nodes.
#> This can be a color name, a hex code, or a column name from the nodes data frame.
#> It has higher priority when it is a column name.
#> - node_color_name: A character string specifying the name of the node color in the legend.
#> - node_shape_by: A numeric value or a character string specifying the column name of the nodes data frame for the node shape.
#> If a numeric value is provided, all nodes will have the same shape.
#> - node_shape_name: A character string specifying the name of the node shape in the legend.
#> - node_fill_by: A character string specifying the fill color of the nodes.
#> This can be a color name, a hex code, or a column name from the nodes data frame.
#> It has higher priority when it is a column name.
#> - node_fill_name: A character string specifying the name of the node fill in the legend.
#> - node_alpha: A numeric value specifying the transparency of the nodes.
#> It only works when the nodes are filled.
#> - node_stroke: A numeric value specifying the stroke of the nodes.
#> - cluster_scale: A character string specifying how to scale the clusters.
#> It can be "fill", "color", or "shape".
#> - node_size_range: A numeric vector specifying the range of the node size.
#> - link_weight_range: A numeric vector specifying the range of the link weight.
#> - link_arrow_offset: A numeric value specifying the offset of the link arrows.
#> So that they won't overlap with the nodes.
#> - link_color_by: A character string specifying the colors of the link. It can be:
#>
#> "from" means the color of the link is determined by the source node.
#> "to" means the color of the link is determined by the target node.
#> Otherwise, the color of the link is determined by the column name from the links data frame.
#>
#> - link_color_name: A character string specifying the name of the link color in the legend.
#> Only used when link_color_by is a column name.
#> - palette: A character string specifying the palette to use.
#> A named list or vector can be used to specify the palettes for different split_by values.
#> - palcolor: A character string specifying the color to use in the palette.
#> A named list can be used to specify the colors for different split_by values.
#> If some values are missing, the values from the palette will be used (palcolor will be NULL for those values).
#> - link_palette: A character string specifying the palette of the links.
#> When link_color_by is "from" or "to", the palette of the links defaults to the palette of the nodes.
#> - link_palcolor: A character vector specifying the colors of the link palette.
#> When link_color_by is "from" or "to", the colors of the link palette defaults to the colors of the node palette.
#> - directed: A logical value specifying whether the graph is directed.
#> - layout: A character string specifying the layout of the graph.
#> It can be "circle", "tree", "grid", or a layout function from igraph.
#> - cluster: A character string specifying the clustering method.
#> It can be "none", "fast_greedy", "walktrap", "edge_betweenness", "infomap", or a clustering function from igraph.
#> - add_mark: A logical value specifying whether to add mark for the clusters to the plot.
#> - mark_expand: A unit value specifying the expansion of the mark.
#> - mark_type: A character string specifying the type of the mark.
#> It can be "hull", "ellipse", "rect", "circle", or a mark function from ggforce.
#> - mark_alpha: A numeric value specifying the transparency of the mark.
#> - mark_linetype: A numeric value specifying the line type of the mark.
#> - add_label: A logical value specifying whether to add label to the nodes to the plot.
#> - label_size: A numeric value specifying the size of the label.
#> - label_fg: A character string specifying the foreground color of the label.
#> - label_bg: A character string specifying the background color of the label.
#> - label_bg_r: A numeric value specifying the background ratio of the label.
#> - arrow: An arrow object for the links.
#> - title: A character string specifying the title of the plot.
#> A function can be used to generate the title based on the default title.
#> This is useful when split_by is used and the title needs to be dynamic.
#> - subtitle: A character string specifying the subtitle of the plot.
#> - xlab: A character string specifying the x-axis label.
#> - ylab: A character string specifying the y-axis label.
#> - aspect.ratio: A numeric value specifying the aspect ratio of the plot.
#> - theme: A character string or a theme class (i.e. ggplot2::theme_classic) specifying the theme to use.
#> Default is "theme_this".
#> - theme_args: A list of arguments to pass to the theme function.
#> - legend.position: A character string specifying the position of the legend.
#> if waiver(), for single groups, the legend will be "none", otherwise "right".
#> - legend.direction: A character string specifying the direction of the legend.
#> - seed: The random seed to use. Default is 8525.
#> - combine: Whether to combine the plots into one when facet is FALSE. Default is TRUE.
#> - nrow: A numeric value specifying the number of rows in the facet.
#> - ncol: A numeric value specifying the number of columns in the facet.
#> - byrow: A logical value indicating whether to fill the plots by row.
#> - axes: A string specifying how axes should be treated. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'keep' will retain all axes in individual plots.
#> 'collect' will remove duplicated axes when placed in the same run of rows or columns of the layout.
#> 'collect_x' and 'collect_y' will remove duplicated x-axes in the columns or duplicated y-axes in the rows respectively.
#>
#> - axis_titles: A string specifying how axis titltes should be treated. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'keep' will retain all axis titles in individual plots.
#> 'collect' will remove duplicated titles in one direction and merge titles in the opposite direction.
#> 'collect_x' and 'collect_y' control this for x-axis titles and y-axis titles respectively.
#>
#> - guides: A string specifying how guides should be treated in the layout. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'collect' will collect guides below to the given nesting level, removing duplicates.
#> 'keep' will stop collection at this level and let guides be placed alongside their plot.
#> 'auto' will allow guides to be collected if a upper level tries, but place them alongside the plot if not.
#>
#> - design: Specification of the location of areas in the layout, passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE. When specified, nrow, ncol, and byrow are ignored.
#> See patchwork::wrap_plots() for more details.
#> For ChordPlot, see plotthis::ChordPlot().
#>
#> [...] can be:
#> - y: A character string specifying the column name of the data frame to plot for the y-axis.
#> - from: A character string of the column name to plot for the source.
#> A character/factor column is expected.
#> - from_sep: A character string to concatenate the columns in from, if multiple columns are provided.
#> - to: A character string of the column name to plot for the target.
#> A character/factor column is expected.
#> - to_sep: A character string to concatenate the columns in to, if multiple columns are provided.
#> - split_by_sep: The separator for multiple split_by columns. See split_by
#> - flip: A logical value to flip the source and target.
#> - links_color: A character string to specify the color of the links.
#> Either "from" or "to".
#> - theme: A character string or a theme class (i.e. ggplot2::theme_classic) specifying the theme to use.
#> Default is "theme_this".
#> - theme_args: A list of arguments to pass to the theme function.
#> - palette: A character string specifying the palette to use.
#> A named list or vector can be used to specify the palettes for different split_by values.
#> - palcolor: A character string specifying the color to use in the palette.
#> A named list can be used to specify the colors for different split_by values.
#> If some values are missing, the values from the palette will be used (palcolor will be NULL for those values).
#> - alpha: A numeric value specifying the transparency of the plot.
#> - labels_rot: A logical value to rotate the labels by 90 degrees.
#> - title: A character string specifying the title of the plot.
#> A function can be used to generate the title based on the default title.
#> This is useful when split_by is used and the title needs to be dynamic.
#> - subtitle: A character string specifying the subtitle of the plot.
#> - seed: The random seed to use. Default is 8525.
#> - combine: Whether to combine the plots into one when facet is FALSE. Default is TRUE.
#> - nrow: A numeric value specifying the number of rows in the facet.
#> - ncol: A numeric value specifying the number of columns in the facet.
#> - byrow: A logical value indicating whether to fill the plots by row.
#> - axes: A string specifying how axes should be treated. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'keep' will retain all axes in individual plots.
#> 'collect' will remove duplicated axes when placed in the same run of rows or columns of the layout.
#> 'collect_x' and 'collect_y' will remove duplicated x-axes in the columns or duplicated y-axes in the rows respectively.
#>
#> - axis_titles: A string specifying how axis titltes should be treated. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'keep' will retain all axis titles in individual plots.
#> 'collect' will remove duplicated titles in one direction and merge titles in the opposite direction.
#> 'collect_x' and 'collect_y' control this for x-axis titles and y-axis titles respectively.
#>
#> - guides: A string specifying how guides should be treated in the layout. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'collect' will collect guides below to the given nesting level, removing duplicates.
#> 'keep' will stop collection at this level and let guides be placed alongside their plot.
#> 'auto' will allow guides to be collected if a upper level tries, but place them alongside the plot if not.
#>
#> - design: Specification of the location of areas in the layout, passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE. When specified, nrow, ncol, and byrow are ignored.
#> See patchwork::wrap_plots() for more details.
#> For Heatmap, see plotthis::Heatmap().
#>
#> [...] can be:
#> - values_by: A character of column name in data that contains the values to be plotted.
#> This is required when in_form is "long". For other formats, the values are pivoted into a column named by values_by.
#> - values_fill: A value to fill in the missing values in the heatmap.
#> When there is missing value in the data, the cluster_rows and cluster_columns will fail.
#> - name: A character string to name the heatmap (will be used to rename values_by).
#> - in_form: The format of the data. Can be one of "matrix", "long", "wide-rows", "wide-columns", or "auto".
#> Defaults to "auto".
#> - split_by_sep: A character string to concat multiple columns in split_by.
#> - rows_by: A vector of column names in data that contains the row information.
#> This is used to create the rows of the heatmap.
#> When in_form is "long" or "wide-columns", this is requied, and multiple columns can be specified,
#> which will be concatenated by rows_by_sep into a single column.
#> - rows_by_sep: A character string to concat multiple columns in rows_by.
#> - rows_split_by: A character of column name in data that contains the split information for rows.
#> - rows_split_by_sep: A character string to concat multiple columns in rows_split_by.
#> - columns_by: A vector of column names in data that contains the column information.
#> This is used to create the columns of the heatmap.
#> When in_form is "long" or "wide-rows", this is required, and multiple columns can be specified,
#> which will be concatenated by columns_by_sep into a single column.
#> - columns_by_sep: A character string to concat multiple columns in columns_by.
#> - columns_split_by: A character of column name in data that contains the split information for columns.
#> - columns_split_by_sep: A character string to concat multiple columns in columns_split_by.
#> - rows_data: A data frame containing additional data for rows, which can be used to add annotations to the heatmap.
#> It will be joined to the main data by rows_by and split_by if split_by exists in rows_data.
#> This is useful for adding additional information to the rows of the heatmap.
#> - columns_data: A data frame containing additional data for columns, which can be used to add annotations to the heatmap.
#> It will be joined to the main data by columns_by and split_by if split_by exists in columns_data.
#> This is useful for adding additional information to the columns of the heatmap.
#> - columns_name: A character string to rename the column created by columns_by, which will be reflected in the name of the annotation or legend.
#> - columns_split_name: A character string to rename the column created by columns_split_by, which will be reflected in the name of the annotation or legend.
#> - rows_name: A character string to rename the column created by rows_by, which will be reflected in the name of the annotation or legend.
#> - rows_split_name: A character string to rename the column created by rows_split_by, which will be reflected in the name of the annotation or legend.
#> - palette: A character string specifying the palette of the heatmap cells.
#> - palcolor: A character vector of colors to override the palette of the heatmap cells.
#> - rows_palette: A character string specifying the palette of the row group annotation.
#> The default is "Paired".
#> - rows_palcolor: A character vector of colors to override the palette of the row group annotation.
#> - rows_split_palette: A character string specifying the palette of the row split annotation.
#> The default is "simspec".
#> - rows_split_palcolor: A character vector of colors to override the palette of the row split annotation.
#> - columns_palette: A character string specifying the palette of the column group annotation.
#> The default is "Paired".
#> - columns_palcolor: A character vector of colors to override the palette of the column group annotation.
#> - columns_split_palette: A character string specifying the palette of the column split annotation.
#> The default is "simspec".
#> - columns_split_palcolor: A character vector of colors to override the palette of the column split annotation.
#> - pie_size_name: A character string specifying the name of the legend for the pie size.
#> - pie_size: A numeric value or a function specifying the size of the pie chart.
#> If it is a function, the function should take count as the argument and return the size.
#> - pie_values: A function or character that can be converted to a function by match.arg()
#> to calculate the values for the pie chart. Default is "length".
#> The function should take a vector of values as the argument and return a single value, for each
#> group in pie_group_by.
#> - pie_name: A character string to rename the column created by pie_group_by, which will be reflected in the name of the annotation or legend.
#> - pie_group_by: A character of column name in data that contains the group information for pie charts.
#> This is used to create pie charts in the heatmap when cell_type is "pie".
#> - pie_group_by_sep: A character string to concat multiple columns in pie_group_by.
#> - pie_palette: A character string specifying the palette of the pie chart.
#> - pie_palcolor: A character vector of colors to override the palette of the pie chart.
#> - bars_sample: An integer specifying the number of samples to draw the bars.
#> - label: A function to calculate the labels for the heatmap cells.
#> It can take either 1, 3, or 5 arguments. The first argument is the aggregated values.
#> If it takes 3 arguments, the second and third arguments are the row and column indices.
#> If it takes 5 arguments, the second and third arguments are the row and column indices,
#> the fourth and fifth arguments are the row and column names.
#> The function should return a character vector of the same length as the aggregated values.
#> If the function returns NA, no label will be shown for that cell.
#> For the indices, if you have the same dimension of data (same order of rows and columns) as the heatmap, you need to use ComplexHeatmap::pindex() to get the correct values.
#> - label_size: A numeric value specifying the size of the labels when cell_type = "label".
#> - violin_fill: A character vector of colors to override the fill color of the violin plot.
#> If NULL, the fill color will be the same as the annotion.
#> - boxplot_fill: A character vector of colors to override the fill color of the boxplot.
#> If NULL, the fill color will be the same as the annotion.
#> - dot_size: A numeric value specifying the size of the dot or a function to calculate the size
#> from the values in the cell or a function to calculate the size from the values in the cell.
#> - dot_size_name: A character string specifying the name of the legend for the dot size.
#> If NULL, the dot size legend will not be shown.
#> - legend_items: A numeric vector with names to specifiy the items in the main legend.
#> The names will be working as the labels of the legend items.
#> - legend_discrete: A logical value indicating whether the main legend is discrete.
#> - legend.position: A character string specifying the position of the legend.
#> if waiver(), for single groups, the legend will be "none", otherwise "right".
#> - legend.direction: A character string specifying the direction of the legend.
#> - lower_quantile, upper_quantile, lower_cutoff, upper_cutoff: Vector of minimum and maximum cutoff values or quantile values for each feature.
#> It's applied to aggregated values when aggregated values are used (e.g. plot_type tile, label, etc).
#> It's applied to raw values when raw values are used (e.g. plot_type bars, etc).
#> - add_bg: A logical value indicating whether to add a background to the heatmap.
#> Does not work with cell_type = "bars" or cell_type = "tile".
#> - bg_alpha: A numeric value between 0 and 1 specifying the transparency of the background.
#> - add_reticle: A logical value indicating whether to add a reticle to the heatmap.
#> - reticle_color: A character string specifying the color of the reticle.
#> - column_name_annotation: A logical value indicating whether to add the column annotation for the column names.
#> which is a simple annotaion indicating the column names.
#> - column_name_legend: A logical value indicating whether to show the legend of the column name annotation.
#> - row_name_annotation: A logical value indicating whether to add the row annotation for the row names.
#> which is a simple annotaion indicating the row names.
#> - row_name_legend: A logical value indicating whether to show the legend of the row name annotation.
#> - cluster_columns: A logical value indicating whether to cluster the columns.
#> If TRUE and columns_split_by is provided, the clustering will only be applied to the columns within the same split.
#> - cluster_rows: A logical value indicating whether to cluster the rows.
#> If TRUE and rows_split_by is provided, the clustering will only be applied to the rows within the same split.
#> - border: A logical value indicating whether to draw the border of the heatmap.
#> If TRUE, the borders of the slices will be also drawn.
#> - title: The global (column) title of the heatmap
#> - column_title: A character string/vector of the column name(s) to use as the title of the column group annotation.
#> - row_title: A character string/vector of the column name(s) to use as the title of the row group annotation.
#> - na_col: A character string specifying the color for missing values.
#> The default is "grey85".
#> - row_names_side: A character string specifying the side of the row names.
#> The default is "right".
#> - column_names_side: A character string specifying the side of the column names.
#> The default is "bottom".
#> - column_annotation: A character string/vector of the column name(s) to use as the column annotation.
#> Or a list with the keys as the names of the annotation and the values as the column names.
#> - column_annotation_side: A character string specifying the side of the column annotation.
#> Could be a list with the keys as the names of the annotation and the values as the sides.
#> - column_annotation_palette: A character string specifying the palette of the column annotation.
#> The default is "Paired".
#> Could be a list with the keys as the names of the annotation and the values as the palettes.
#> - column_annotation_palcolor: A character vector of colors to override the palette of the column annotation.
#> Could be a list with the keys as the names of the annotation and the values as the palcolors.
#> - column_annotation_type: A character string specifying the type of the column annotation.
#> The default is "auto". Other options are "simple", "pie", "ring", "bar", "violin", "boxplot", "density".
#> Could be a list with the keys as the names of the annotation and the values as the types.
#> If the type is "auto", the type will be determined by the type and number of the column data.
#> - column_annotation_params: A list of parameters passed to the annotation function.
#> Could be a list with the keys as the names of the annotation and the values as the parameters passed to the annotation function. For the parameters for names (columns_by, rows_by, columns_split_by, rows_split_by), the key should be "name.(name)", where (name) is the name of the annotation.
#> See anno_pie(), anno_ring(), anno_bar(), anno_violin(), anno_boxplot(), anno_density(), anno_simple(), anno_points() and anno_lines() for the parameters of each annotation function.
#> - column_annotation_agg: A function to aggregate the values in the column annotation.
#> - row_annotation: A character string/vector of the column name(s) to use as the row annotation.
#> Or a list with the keys as the names of the annotation and the values as the column names.
#> - row_annotation_side: A character string specifying the side of the row annotation.
#> Could be a list with the keys as the names of the annotation and the values as the sides.
#> - row_annotation_palette: A character string specifying the palette of the row annotation.
#> The default is "Paired".
#> Could be a list with the keys as the names of the annotation and the values as the palettes.
#> - row_annotation_palcolor: A character vector of colors to override the palette of the row annotation.
#> Could be a list with the keys as the names of the annotation and the values as the palcolors.
#> - row_annotation_type: A character string specifying the type of the row annotation.
#> The default is "auto". Other options are "simple", "pie", "ring", "bar", "violin", "boxplot", "density".
#> Could be a list with the keys as the names of the annotation and the values as the types.
#> If the type is "auto", the type will be determined by the type and number of the row data.
#> - row_annotation_params: A list of parameters passed to the annotation function.
#> Could be a list with the keys as the names of the annotation and the values as the parameters.
#> Same as column_annotation_params.
#> - row_annotation_agg: A function to aggregate the values in the row annotation.
#> - flip: A logical value indicating whether to flip the heatmap.
#> The idea is that, you can simply set flip = TRUE to flip the heatmap.
#> You don't need to swap the arguments related to rows and columns, except those you specify via ...
#> that are passed to ComplexHeatmap::Heatmap() directly.
#> - alpha: A numeric value between 0 and 1 specifying the transparency of the heatmap cells.
#> - seed: The random seed to use. Default is 8525.
#> - layer_fun_callback: A function to add additional layers to the heatmap.
#> The function should have the following arguments: j, i, x, y, w, h, fill, sr and sc.
#> Please also refer to the layer_fun argument in ComplexHeatmap::Heatmap.
#> - cell_type: A character string specifying the type of the heatmap cells.
#> The default is values. Other options are "bars", "label", "dot", "violin", "boxplot".
#> Note that for pie chart, the values under columns specified by rows will not be used directly. Instead, the values
#> will just be counted in different pie_group_by groups. NA values will not be counted.
#> - cell_agg: A function to aggregate the values in the cell, for the cell type "tile" and "label".
#> The default is mean.
#> - combine: Whether to combine the plots into one when facet is FALSE. Default is TRUE.
#> - nrow: A numeric value specifying the number of rows in the facet.
#> - ncol: A numeric value specifying the number of columns in the facet.
#> - byrow: A logical value indicating whether to fill the plots by row.
#> - axes: A string specifying how axes should be treated. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'keep' will retain all axes in individual plots.
#> 'collect' will remove duplicated axes when placed in the same run of rows or columns of the layout.
#> 'collect_x' and 'collect_y' will remove duplicated x-axes in the columns or duplicated y-axes in the rows respectively.
#>
#> - axis_titles: A string specifying how axis titltes should be treated. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'keep' will retain all axis titles in individual plots.
#> 'collect' will remove duplicated titles in one direction and merge titles in the opposite direction.
#> 'collect_x' and 'collect_y' control this for x-axis titles and y-axis titles respectively.
#>
#> - guides: A string specifying how guides should be treated in the layout. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'collect' will collect guides below to the given nesting level, removing duplicates.
#> 'keep' will stop collection at this level and let guides be placed alongside their plot.
#> 'auto' will allow guides to be collected if a upper level tries, but place them alongside the plot if not.
#>
#> - design: Specification of the location of areas in the layout, passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE. When specified, nrow, ncol, and byrow are ignored.
#> See patchwork::wrap_plots() for more details.
#> For SankeyPlot, see plotthis::SankeyPlot().
#>
#> [...] can be:
#> - in_form: A character string to specify the format of the data.
#> Possible values are "auto", "long", "lodes", "wide", "alluvia", and "counts".
#> - x: A character string specifying the column name of the data frame to plot for the x-axis.
#> - x_sep: A character string to concatenate the columns in x, if multiple columns are provided.
#> - y: A character string specifying the column name of the data frame to plot for the y-axis.
#> - stratum: A character string of the column name to group the nodes for each x.
#> See data for more details.
#> - stratum_sep: A character string to concatenate the columns in stratum, if multiple columns are provided.
#> - alluvium: A character string of the column name to define the links.
#> See data for more details.
#> - alluvium_sep: A character string to concatenate the columns in alluvium, if multiple columns are provided.
#> - split_by_sep: The separator for multiple split_by columns. See split_by
#> - keep_empty: A logical value indicating whether to keep empty groups.
#> If FALSE, empty groups will be removed.
#> - flow: A logical value to use ggalluvial::geom_flow instead of ggalluvial::geom_alluvium.
#> - expand: The values to expand the x and y axes. It is like CSS padding.
#> When a single value is provided, it is used for both axes on both sides.
#> When two values are provided, the first value is used for the top/bottom side and the second value is used for the left/right side.
#> When three values are provided, the first value is used for the top side, the second value is used for the left/right side, and the third value is used for the bottom side.
#> When four values are provided, the values are used for the top, right, bottom, and left sides, respectively.
#> You can also use a named vector to specify the values for each side.
#> When the axis is discrete, the values will be applied as 'add' to the 'expansion' function.
#> When the axis is continuous, the values will be applied as 'mult' to the 'expansion' function.
#> See also https://ggplot2.tidyverse.org/reference/expansion.html
#> - nodes_legend: Controls how the legend of nodes will be shown. Possible values are:
#>
#> "merge": Merge the legends of nodes. That is only one legend will be shown for all nodes.
#> "separate": Show the legends of nodes separately. That is, nodes on each x will have their own legend.
#> "none": Do not show the legend of nodes.
#> "auto": Automatically determine how to show the legend.
#> When nodes_label is TRUE, "none" will apply.
#> When nodes_label is FALSE, and if stratum is the same as links_fill_by, "none" will apply.
#> If there is any overlapping values between the nodes on different x,
#> "merge" will apply. Otherwise, "separate" will apply.
#>
#> - nodes_color: A character string to color the nodes.
#> Use a special value ".fill" to use the same color as the fill.
#> - links_fill_by: A character string of the column name to fill the links.
#> - links_fill_by_sep: A character string to concatenate the columns in links_fill_by, if multiple columns are provided.
#> - links_name: A character string to name the legend of links.
#> - links_color: A character string to color the borders of links.
#> Use a special value ".fill" to use the same color as the fill.
#> - nodes_palette: A character string to specify the palette of nodes fill.
#> - nodes_palcolor: A character vector to specify the colors of nodes fill.
#> - nodes_alpha: A numeric value to specify the transparency of nodes fill.
#> - nodes_label: A logical value to show the labels on the nodes.
#> - nodes_label_miny: A numeric value to specify the minimum y (frequency) to show the labels.
#> - nodes_width: A numeric value to specify the width of nodes.
#> - links_palette: A character string to specify the palette of links fill.
#> - links_palcolor: A character vector to specify the colors of links fill.
#> - links_alpha: A numeric value to specify the transparency of links fill.
#> - legend.box: A character string to specify the box of the legend, either "vertical" or "horizontal".
#> - aspect.ratio: A numeric value specifying the aspect ratio of the plot.
#> - legend.position: A character string specifying the position of the legend.
#> if waiver(), for single groups, the legend will be "none", otherwise "right".
#> - legend.direction: A character string specifying the direction of the legend.
#> - flip: A logical value to flip the plot.
#> - theme: A character string or a theme class (i.e. ggplot2::theme_classic) specifying the theme to use.
#> Default is "theme_this".
#> - theme_args: A list of arguments to pass to the theme function.
#> - title: A character string specifying the title of the plot.
#> A function can be used to generate the title based on the default title.
#> This is useful when split_by is used and the title needs to be dynamic.
#> - subtitle: A character string specifying the subtitle of the plot.
#> - xlab: A character string specifying the x-axis label.
#> - ylab: A character string specifying the y-axis label.
#> - facet_scales: Whether to scale the axes of facets. Default is "fixed"
#> Other options are "free", "free_x", "free_y". See ggplot2::facet_wrap
#> - facet_ncol: A numeric value specifying the number of columns in the facet.
#> When facet_by is a single column and facet_wrap is used.
#> - facet_nrow: A numeric value specifying the number of rows in the facet.
#> When facet_by is a single column and facet_wrap is used.
#> - facet_byrow: A logical value indicating whether to fill the plots by row. Default is TRUE.
#> - seed: The random seed to use. Default is 8525.
#> - combine: Whether to combine the plots into one when facet is FALSE. Default is TRUE.
#> - nrow: A numeric value specifying the number of rows in the facet.
#> - ncol: A numeric value specifying the number of columns in the facet.
#> - byrow: A logical value indicating whether to fill the plots by row.
#> - axes: A string specifying how axes should be treated. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'keep' will retain all axes in individual plots.
#> 'collect' will remove duplicated axes when placed in the same run of rows or columns of the layout.
#> 'collect_x' and 'collect_y' will remove duplicated x-axes in the columns or duplicated y-axes in the rows respectively.
#>
#> - axis_titles: A string specifying how axis titltes should be treated. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'keep' will retain all axis titles in individual plots.
#> 'collect' will remove duplicated titles in one direction and merge titles in the opposite direction.
#> 'collect_x' and 'collect_y' control this for x-axis titles and y-axis titles respectively.
#>
#> - guides: A string specifying how guides should be treated in the layout. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'collect' will collect guides below to the given nesting level, removing duplicates.
#> 'keep' will stop collection at this level and let guides be placed alongside their plot.
#> 'auto' will allow guides to be collected if a upper level tries, but place them alongside the plot if not.
#>
#> - design: Specification of the location of areas in the layout, passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE. When specified, nrow, ncol, and byrow are ignored.
#> See patchwork::wrap_plots() for more details.
#> For DotPlot, see plotthis::DotPlot().
#>
#> [...] can be:
#> - x: A character vector specifying the column to use for the x-axis.
#> A numeric column is expected.
#> - y: A character vector specifying the column to use for the y-axis.
#> A factor/character column is expected.
#> - x_sep: A character vector to concatenate multiple columns in x. Default is "_".
#> - y_sep: A character vector to concatenate multiple columns in y. Default is "_".
#> - flip: A logical value indicating whether to flip the x and y axes. Default is FALSE.
#> - split_by_sep: The separator for multiple split_by columns. See split_by
#> - size_name: A character vector specifying the name for the size legend.
#> - fill_name: A character vector specifying the name for the fill legend.
#> - fill_cutoff_name: A character vector specifying the name for the fill cutoff legend.
#> - add_bg: A logical value indicating whether to add a background color to the plot. Default is FALSE.
#> - bg_palette: A character vector specifying the palette for the background color. Default is "stripe".
#> - bg_palcolor: A character vector specifying the color for the background color.
#> - bg_alpha: A numeric value specifying the alpha for the background color. Default is 0.2.
#> - bg_direction: A character vector specifying the direction for the background color. Default is "vertical".
#> Other options are "horizontal". "h" and "v" are also accepted.
#> - size_by: Which column to use as the size of the dots. It must be a numeric column.
#> If not provided, the size will be the count of the instances for each 'y' in 'x'.
#> For 'ScatterPlot', it can be a single numeric value to specify the size of the dots.
#> - fill_by: Which column to use as the fill the dots. It must be a numeric column.
#> If not provided, all dots will be filled with the same color at the middle of the palette.
#> - fill_cutoff: A numeric value specifying the cutoff for the fill column.
#> - fill_reverse: A logical value indicating whether to reverse the fill direction. Default is FALSE.
#> By default, the fill direction is "up". If TRUE, the fill direction is "down".
#> When the direction is "up", the values less than the cutoff will be filled with grey.
#> When the direction is "down", the values greater than the cutoff will be filled with grey.
#> - theme: A character string or a theme class (i.e. ggplot2::theme_classic) specifying the theme to use.
#> Default is "theme_this".
#> - theme_args: A list of arguments to pass to the theme function.
#> - palette: A character string specifying the palette to use.
#> A named list or vector can be used to specify the palettes for different split_by values.
#> - palcolor: A character string specifying the color to use in the palette.
#> A named list can be used to specify the colors for different split_by values.
#> If some values are missing, the values from the palette will be used (palcolor will be NULL for those values).
#> - alpha: A numeric value specifying the transparency of the plot.
#> - facet_scales: Whether to scale the axes of facets. Default is "fixed"
#> Other options are "free", "free_x", "free_y". See ggplot2::facet_wrap
#> - facet_ncol: A numeric value specifying the number of columns in the facet.
#> When facet_by is a single column and facet_wrap is used.
#> - facet_nrow: A numeric value specifying the number of rows in the facet.
#> When facet_by is a single column and facet_wrap is used.
#> - facet_byrow: A logical value indicating whether to fill the plots by row. Default is TRUE.
#> - seed: The random seed to use. Default is 8525.
#> - aspect.ratio: A numeric value specifying the aspect ratio of the plot.
#> - legend.position: A character string specifying the position of the legend.
#> if waiver(), for single groups, the legend will be "none", otherwise "right".
#> - legend.direction: A character string specifying the direction of the legend.
#> - title: A character string specifying the title of the plot.
#> A function can be used to generate the title based on the default title.
#> This is useful when split_by is used and the title needs to be dynamic.
#> - subtitle: A character string specifying the subtitle of the plot.
#> - xlab: A character string specifying the x-axis label.
#> - ylab: A character string specifying the y-axis label.
#> - keep_empty: A logical value indicating whether to keep empty groups.
#> If FALSE, empty groups will be removed.
#> - combine: Whether to combine the plots into one when facet is FALSE. Default is TRUE.
#> - nrow: A numeric value specifying the number of rows in the facet.
#> - ncol: A numeric value specifying the number of columns in the facet.
#> - byrow: A logical value indicating whether to fill the plots by row.
#> - axes: A string specifying how axes should be treated. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'keep' will retain all axes in individual plots.
#> 'collect' will remove duplicated axes when placed in the same run of rows or columns of the layout.
#> 'collect_x' and 'collect_y' will remove duplicated x-axes in the columns or duplicated y-axes in the rows respectively.
#>
#> - axis_titles: A string specifying how axis titltes should be treated. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'keep' will retain all axis titles in individual plots.
#> 'collect' will remove duplicated titles in one direction and merge titles in the opposite direction.
#> 'collect_x' and 'collect_y' control this for x-axis titles and y-axis titles respectively.
#>
#> - guides: A string specifying how guides should be treated in the layout. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'collect' will collect guides below to the given nesting level, removing duplicates.
#> 'keep' will stop collection at this level and let guides be placed alongside their plot.
#> 'auto' will allow guides to be collected if a upper level tries, but place them alongside the plot if not.
#>
#> - design: Specification of the location of areas in the layout, passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE. When specified, nrow, ncol, and byrow are ignored.
#> See patchwork::wrap_plots() for more details.
#>
#> - value
#>
#> A ggplot object or a list if combine is FALSE
#>
#> - examples
#>
#>
#> set.seed(8525)
#> data(cellphonedb_res)
#> CCCPlot(data = cellphonedb_res, plot_type = "network", legend.position = "none",
#> theme = "theme_blank", theme_args = list(add_coord = FALSE))
#> CCCPlot(cellphonedb_res, plot_type = "chord")
#> CCCPlot(cellphonedb_res, plot_type = "heatmap")
#> CCCPlot(cellphonedb_res, plot_type = "dot",
#> magnitude_agg = mean, magnitude_name = "Average Interaction Strength")
#> CCCPlot(cellphonedb_res, plot_type = "sankey")
#>
#> cellphonedb_res_sub <- cellphonedb_res[
#> cellphonedb_res$source %in% c("Dendritic", "CD14+ Monocyte"),]
#> CCCPlot(cellphonedb_res_sub, plot_type = "dot", method = "interaction")
#> CCCPlot(cellphonedb_res_sub, plot_type = "network", method = "interaction",
#> node_size_by = 1)
#> CCCPlot(cellphonedb_res_sub, plot_type = "heatmap", method = "interaction",
#> palette = "Reds")
#> CCCPlot(cellphonedb_res_sub, plot_type = "box", method = "interaction")
#> CCCPlot(cellphonedb_res_sub, plot_type = "violin", method = "interaction",
#> add_box = TRUE)
#> CCCPlot(cellphonedb_res_sub, plot_type = "ridge", method = "interaction")
#>
#>
#>
#>
#> You must code in the programming language 'R' to answer this prompt.
#> You can use functions from these packages: scplotter.
#> You may not install or load any additional packages.
#> These objects already exist in the R session:
#>
#> Object_name, Type
#> cellphonedb_res, data.frame.
#>
#> Do not define these objects in your R code.#> --- Receiving response from LLM provider: ---#> ```r
#> CCCPlot(cellphonedb_res, plot_type = "dot")
#> ```#> Code ran:
#> CCCPlot(cellphonedb_res, plot_type = "dot")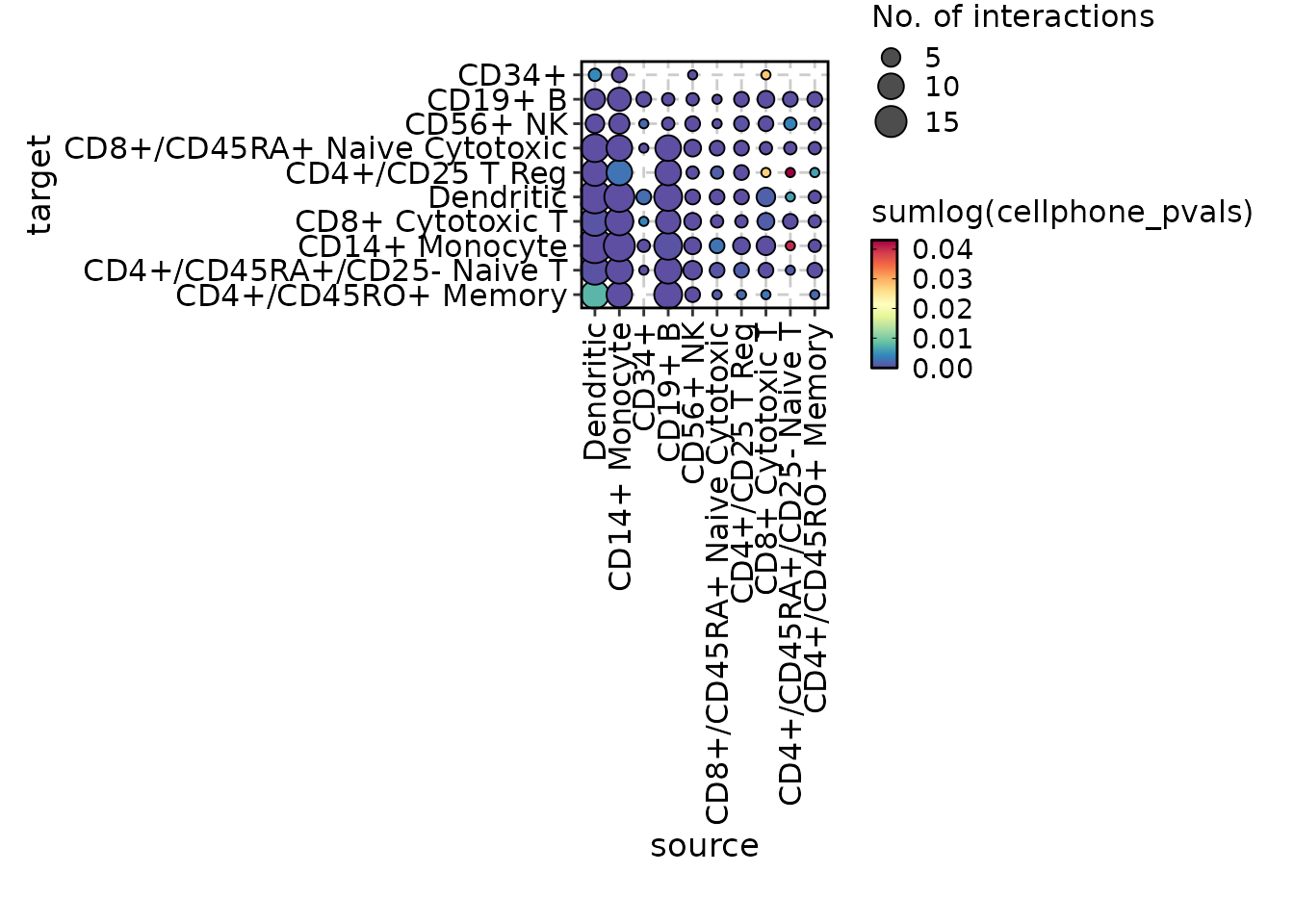
To only debug a single conversation, you can set verbose
to TRUE in the ask() method.
chat <- SCPlotterChat$new(
provider = provider,
verbose = FALSE
)
chat$ask("Generate a cell-cell communication plot for the cellphonedb_res data.", verbose = TRUE)
#> --- Sending request to LLM provider (gpt-5-nano): ---
#> Objective: Select the most appropriate tool to handle the user's request while preserving conversational context. If the user refines or changes how the previous result should be visualized (e.g., asks for a different plot type), continue with the last plotting tool used unless they explicitly name a different tool.
#>
#> Decision Process:
#> - If the user explicitly names a tool, output that tool.
#> - Else, if the request appears to refine the previous output (e.g., "do X instead", "make it a heatmap/dot/bar/etc", "change to ...", "add ... to", "same plot but ..."), select the last tool mentioned in the chat history.
#> - Else, analyze the current user request; if there is a clear, unambiguous match to a single tool, select that tool.
#> - Else, use the last mentioned tool from the chat history.
#> - If no tool is found, respond with "None".
#>
#> Response Format: Provide only the name of the selected tool, or "None" if no tool applies.
#>
#> User Request:
#> Generate a cell-cell communication plot for the cellphonedb_res data.
#>
#> Available Tools:
#> - gt: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - CellDimPlot: Cell Dimension Reduction Plot
#> This function creates a dimension reduction plot for a Seurat object
#> a Giotto object, a path to an .h5ad file or an opened H5File by hdf5r package.
#> It allows for various customizations such as grouping by metadata,
#> adding edges between cell neighbors, highlighting specific cells, and more.
#> This function is a wrapper around plotthis::DimPlot() , which provides a
#> flexible way to visualize cell clusters in reduced dimensions. This function
#> extracts the necessary data from the Seurat or Giotto object and passes it to
#> plotthis::DimPlot() .
#>
#> - ClonalVolumePlot: ClonalVolumePlot
#> ClonalVolumePlot
#>
#> - ClonalRarefactionPlot: ClonalRarefactionPlot
#> Plot the rarefaction curves
#>
#> - ClonalStatPlot: ClonalStatPlot
#> Visualize the statistics of the clones.
#>
#> - ClonalCompositionPlot: ClonalCompositionPlot
#> Plot the composition of the clones in different samples/groups.
#>
#> - EnrichmentPlot: Enrichment Plot
#> This function generates various types of plots for enrichment (over-representation) analysis.
#>
#> - eq: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - CellVelocityPlot: Cell Velocity Plot
#> This function creates a cell velocity plot for a Seurat object,
#> a Giotto object, a path to an .h5ad file or an opened H5File by hdf5r package.
#> It allows for various customizations such as grouping by metadata,
#> adding edges between cell neighbors, highlighting specific cells, and more.
#> This function is a wrapper around plotthis::VelocityPlot() , which provides a
#> flexible way to visualize cell velocities in reduced dimensions. This function
#> extracts the cell embeddings and velocity embeddings from the Seurat or Giotto object
#> and passes them to plotthis::VelocityPlot() .
#>
#> - SpatFeaturePlot: Plot features for spatial data
#> The features can include expression, dimension reduction components, metadata, etc
#>
#> - shared: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - CellStatPlot: Cell statistics plot
#> This function creates a plot to visualize the statistics of cells in a Seurat object, a Giotto object,
#> a path to an .h5ad file or an opened H5File by hdf5r package.
#> It can create various types of plots, including bar plots, circos plots, pie charts, pies (heatmap with cell_type = 'pie'), ring/donut plots, trend plots
#> area plots, sankey/alluvial plots, heatmaps, radar plots, spider plots, violin plots, and box plots.
#> The function allows for grouping, splitting, and faceting the data based on metadata columns.
#> It also supports calculating fractions of cells based on specified groupings.#'
#>
#> - ne: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - SpatDimPlot: Plot categories for spatial data
#> Plot categories for spatial data
#>
#> - GSEASummaryPlot: Objects exported from other packages
#> These objects are imported from other packages. Follow the links
#> below to see their documentation.
#>
#>
#> plotthis GSEAPlot , GSEASummaryPlot
#>
#> - CCCPlot: Cell-Cell Communication Plot
#> Plot the cell-cell communication.
#> See also:
#>
#> The review: https://www.sciencedirect.com/science/article/pii/S2452310021000081
#> The LIANA package: https://liana-py.readthedocs.io/en/latest/notebooks/basic_usage.html#Tileplot
#> The CCPlotR package: https://github.com/Sarah145/CCPlotR
#>
#>
#> - ClonalResidencyPlot: ClonalResidencyPlot
#> Plot the residency of the clones in different samples.
#>
#> - ClustreePlot: Clustree plot
#> This function generates a clustree plot from a data frame or a Seurat object.
#>
#> - ClonalKmerPlot: ClonalKmerPlot
#> Explore the k-mer frequency of CDR3 sequences.
#>
#> - ClonalPositionalPlot: ClonalPositionalPlot
#> Visualize the positional entropy, property or amino acid frequency of CDR3 sequences.
#>
#> - uniq: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - top: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - le: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - MarkersPlot: Visualize Markers
#> Plot markers, typically identified by Seurat::FindMarkers() or Seurat::FindAllMarkers() .
#>
#> - ClonalOverlapPlot: ClonalOverlapPlot
#> Plot the overlap of the clones in different samples/groups.
#>
#> - ge: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - ClonalDynamicsPlot: ClonalDynamicsPlot
#> This function is deprecated. Please use ClonalStatPlot() instead.
#>
#> - ClonalLengthPlot: ClonalLengthPlot
#> Plot the length distribution of the CDR3 sequences
#>
#> - and: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - ClonalAbundancePlot: ClonalAbundancePlot
#> Plot the count or density of the clones at different abundance levels.
#>
#> - sel: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - ClonalDiversityPlot: ClonalDiversityPlot
#> Plot the clonal diversities of the samples/groups.
#>
#> - FeatureStatPlot: Feature statistic plot
#> This function creates various types of feature statistic plots for a Seurat object, a Giotto object,
#> a path to an .h5ad file or an opened H5File by hdf5r package.
#> It allows for plotting features such as gene expression, scores, or other metadata across different groups or conditions.
#> The function supports multiple plot types including violin, box, bar, ridge, dimension reduction, correlation, heatmap, and dot plots.
#> It can also handle multiple features and supports faceting, splitting, and grouping by metadata columns.
#>
#> - ClonalGeneUsagePlot: ClonalGeneUsagePlot
#> ClonalGeneUsagePlot
#>
#> - lt: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - GSEAPlot: Objects exported from other packages
#> These objects are imported from other packages. Follow the links
#> below to see their documentation.
#>
#>
#> plotthis GSEAPlot , GSEASummaryPlot
#>
#> - or: Helper functions to select clones based on various criteria
#> These helper functions allow for the selection of clones based on various criteria such as size, group comparison, and existence in specific groups.
#>
#> - ListTools: List all available tools
#> List all available tools that can be used to handle the chat request.
#>
#> - ListData: List all available data objects
#> List all available data objects that can be used to handle the chat request.
#> --- Receiving response from LLM provider: ---
#> CCCPlot
#> Tool identified: CCCPlot
#> --- Sending request to LLM provider (gpt-5-nano): ---
#> Objective: Identify the data object to be used.
#>
#> Decision Process:
#> - If the user explicitly names a data object, output that data object name. If the name does not match exactly the available ones, use the one that is mostly related.
#> - Else, if the request appears to refine the previous output (e.g., "do X instead", "make it a heatmap/dot/bar/etc", "change to ...", "add ... to", "same plot but ..."), select the last dataset name mentioned in the chat history.
#> - Else, analyze the current user request; if there is a clear, unambiguous match to a dataset, select that data object.
#> - Else, use the last mentioned data object from the chat history.
#> - If no data object is found, respond with "None".
#>
#> Response Format: Provide only the name of the selected data object, or "None" if no tool applies. The response should be unquoted.
#>
#> User Request:
#> Generate a cell-cell communication plot for the cellphonedb_res data.
#>
#> Available Data Objects:
#> - scplotter::cellphonedb_res: A toy example of CellPhoneDB output from LIANA
#> - scplotter::ifnb_sub: A subsetted version of 'ifnb' datasets
#> - scplotter::pancreas_sub: A subsetted version of mouse 'pancreas' datasets
#> - Seurat::cc.genes: Cell cycle genes
#> - Seurat::cc.genes.updated.2019: Cell cycle genes: 2019 update
#> - SeuratObject::pbmc_small: A small example version of the PBMC dataset
#> - scRepertoire::contig_list: A list of 8 single-cell T cell receptor sequences runs.
#> - scRepertoire::mini_contig_list: Processed subset of 'contig_list'
#> - scRepertoire::scRep_example: A Seurat object of 500 single T cells,
#> --- Receiving response from LLM provider: ---
#> scplotter::cellphonedb_res
#> Data object identified: scplotter::cellphonedb_res
#> Warning in wrap$modify_fn(prompt_text, llm_provider): The 'skimr' package is
#> required to skim dataframes. Skim summary of dataframes currently not shown in
#> prompt
#> --- Sending request to LLM provider (gpt-5-nano): ---#> Objective: Generate the R code to run the specified tool using the specified data object based on the user's request and the provided tool information.
#>
#> Decision Process:
#> - Analyze the user's request to understand what needs to be done with the specified tool and data object.
#> - When use the data object name, do not quote it.
#> - Refer to the provided tool information to understand the tool's usage, arguments, and examples.
#> - Consider any specific parameters or options mentioned in the user's request that need to be included in the tool call.
#> - If the user's request is ambiguous or lacks necessary details, refer to the chat history for additional context that may help clarify the intended use of the tool and data object.
#> - Construct the R code that correctly calls the tool with the specified data object, ensuring that all necessary arguments are included and correctly formatted.
#>
#> Response Format: Provide only the valid R code wrapped between "```r" and "```" to run the tool.
#>
#> Tool to be used: CCCPlot
#> Data object to be used: cellphonedb_res
#>
#> User Request:
#> Generate a cell-cell communication plot for the cellphonedb_res data.
#>
#> Tool Information:
#> - title
#> Cell-Cell Communication Plot
#> - description
#>
#> Plot the cell-cell communication.
#> See also:
#>
#> The review: https://www.sciencedirect.com/science/article/pii/S2452310021000081
#> The LIANA package: https://liana-py.readthedocs.io/en/latest/notebooks/basic_usage.html#Tileplot
#> The CCPlotR package: https://github.com/Sarah145/CCPlotR
#>
#>
#> - usage
#>
#> CCCPlot(
#> data,
#> plot_type = c("dot", "network", "chord", "circos", "heatmap", "sankey", "alluvial",
#> "box", "violin", "ridge"),
#> method = c("aggregation", "interaction"),
#> magnitude = waiver(),
#> specificity = waiver(),
#> magnitude_agg = length,
#> magnitude_name = "No. of interactions",
#> meta_specificity = "sumlog",
#> split_by = NULL,
#> x_text_angle = 90,
#> link_curvature = 0.2,
#> link_alpha = 0.6,
#> facet_by = NULL,
#> show_row_names = TRUE,
#> show_column_names = TRUE,
#> ...
#> )
#>
#> - arguments
#> - data: A data frame with the cell-cell communication data.
#> A typical data frame should have the following columns:
#>
#> source The source cell type.
#> target The target cell type.
#> ligand The ligand gene.
#> receptor The receptor gene.
#> ligand_means The mean expression of the ligand gene per cell type.
#> receptor_means The mean expression of the receptor gene per cell type.
#> ligand_props The proportion of cells that express the entity.
#> receptor_props The proportion of cells that express the entity.
#> <magnitude> The magnitude of the communication.
#> <specificity> The specificity of the communication.
#> Depends on the plot_type, some columns are optional. But the source, target,
#> ligand, receptor and <magnitude> are required.
#>
#> - plot_type: The type of plot to use. Default is "dot".
#> Possible values are "network", "chord", "circos", "heatmap", "sankey", "alluvial", "dot",
#> "box", "violin" and "ridge".
#> For "box", "violin" and "ridge", the method should be "interaction".
#>
#> network: A network plot with the source and target cells as the nodes and the communication as the edges.
#> chord: A chord plot with the source and target cells as the nodes and the communication as the chords.
#> circos: Alias of "chord".
#> heatmap: A heatmap plot with the source and target cells as the rows and columns.
#> sankey: A sankey plot with the source and target cells as the nodes and the communication as the flows.
#> alluvial: Alias of "sankey".
#> dot: A dot plot with the source and target cells as the nodes and the communication as the dots.
#> box: Box plots for source cell types. Each x is a target cell type and the values will be
#> the interaction strengths of the ligand-receptor pairs.
#> violin: Violin plots for source cell types. Each x is a target cell type and the values will be
#> the interaction strengths of the ligand-receptor pairs.
#> ridge: Ridge plots for source cell types. Each row is a target cell type and the values will be
#> the interaction strengths of the ligand-receptor pairs.
#>
#> - method: The method to determine the plot entities.
#>
#> aggregation: Aggregate the ligand-receptor pairs interactions for each source-target pair.
#> Only the source / target pairs will be plotted.
#> interaction: Plot the ligand-receptor pairs interactions directly.
#> The ligand-receptor pairs will also be plotted.
#>
#> - magnitude: The column name in the data to use as the magnitude of the communication.
#> By default, the second last column will be used.
#> See li.mt.show_methods() for the available methods in LIANA.
#> or https://liana-py.readthedocs.io/en/latest/notebooks/basic_usage.html#Tileplot
#> - specificity: The column name in the data to use as the specificity of the communication.
#> By default, the last column will be used.
#> If the method doesn't have a specificity, set it to NULL.
#> - magnitude_agg: A function to aggregate the magnitude of the communication.
#> Default is length.
#> - magnitude_name: The name of the magnitude in the plot.
#> Default is "No. of interactions".
#> - meta_specificity: The method to calculate the specificity when there are multiple
#> ligand-receptor pairs interactions. Default is "sumlog".
#> It should be one of the methods in the metap package.
#> Current available methods are:
#>
#> invchisq: Combine p values using the inverse chi squared method
#> invt: Combine p values using the inverse t method
#> logitp: Combine p values using the logit method
#> meanp: Combine p values by the mean p method
#> meanz: Combine p values using the mean z method
#> sumlog: Combine p-values by the sum of logs (Fisher's) method
#> sump: Combine p-values using the sum of p (Edgington's) method
#> two2one: Convert two-sided p-values to one-sided
#> votep: Combine p-values by the vote counting method
#> wilkinsonp: Combine p-values using Wilkinson's method
#>
#> - split_by: A character vector of column names to split the plots. Default is NULL.
#> - x_text_angle: The angle of the x-axis text. Default is 90.
#> Only used when plot_type is "dot".
#> - link_curvature: The curvature of the links. Default is 0.2.
#> Only used when plot_type is "network".
#> - link_alpha: The transparency of the links. Default is 0.6.
#> Only used when plot_type is "network".
#> - facet_by: A character vector of column names to facet the plots. Default is NULL.
#> It should always be NULL.
#> - show_row_names: Whether to show the row names in the heatmap. Default is TRUE.
#> Only used when plot_type is "heatmap".
#> - show_column_names: Whether to show the column names in the heatmap. Default is TRUE.
#> Only used when plot_type is "heatmap".
#> - ...: Other arguments passed to the specific plot function.
#>
#> For Network, see plotthis::Network().
#>
#> [...] can be:
#> - links: A data frame containing the links between nodes.
#> - nodes: A data frame containing the nodes.
#> This is optional. The names of the nodes are extracted from the links data frame.
#> If "@nodes" is provided, the nodes data frame will be extracted from the attribute nodes of the links data frame.
#> - split_by_sep: The separator for multiple split_by columns. See split_by
#> - split_nodes: A logical value specifying whether to split the nodes data.
#> If TRUE, the nodes data will also be split by the split_by column.
#> - from: A character string specifying the column name of the links data frame for the source nodes.
#> Default is the first column of the links data frame.
#> - from_sep: A character string to concatenate the columns in from, if multiple columns are provided.
#> - to: A character string specifying the column name of the links data frame for the target nodes.
#> Default is the second column of the links data frame.
#> - to_sep: A character string to concatenate the columns in to, if multiple columns are provided.
#> - node_by: A character string specifying the column name of the nodes data frame for the node names.
#> Default is the first column of the nodes data frame.
#> - node_by_sep: A character string to concatenate the columns in node_by, if multiple columns are provided.
#> - link_weight_by: A numeric value or a character string specifying the column name of the links data frame for the link weight.
#> If a numeric value is provided, all links will have the same weight.
#> This determines the width of the links.
#> - link_weight_name: A character string specifying the name of the link weight in the legend.
#> - link_type_by: A character string specifying the type of the links.
#> This can be "solid", "dashed", "dotted", or a column name from the links data frame.
#> It has higher priority when it is a column name.
#> - link_type_name: A character string specifying the name of the link type in the legend.
#> - node_size_by: A numeric value or a character string specifying the column name of the nodes data frame for the node size.
#> If a numeric value is provided, all nodes will have the same size.
#> - node_size_name: A character string specifying the name of the node size in the legend.
#> - node_color_by: A character string specifying the color of the nodes.
#> This can be a color name, a hex code, or a column name from the nodes data frame.
#> It has higher priority when it is a column name.
#> - node_color_name: A character string specifying the name of the node color in the legend.
#> - node_shape_by: A numeric value or a character string specifying the column name of the nodes data frame for the node shape.
#> If a numeric value is provided, all nodes will have the same shape.
#> - node_shape_name: A character string specifying the name of the node shape in the legend.
#> - node_fill_by: A character string specifying the fill color of the nodes.
#> This can be a color name, a hex code, or a column name from the nodes data frame.
#> It has higher priority when it is a column name.
#> - node_fill_name: A character string specifying the name of the node fill in the legend.
#> - node_alpha: A numeric value specifying the transparency of the nodes.
#> It only works when the nodes are filled.
#> - node_stroke: A numeric value specifying the stroke of the nodes.
#> - cluster_scale: A character string specifying how to scale the clusters.
#> It can be "fill", "color", or "shape".
#> - node_size_range: A numeric vector specifying the range of the node size.
#> - link_weight_range: A numeric vector specifying the range of the link weight.
#> - link_arrow_offset: A numeric value specifying the offset of the link arrows.
#> So that they won't overlap with the nodes.
#> - link_color_by: A character string specifying the colors of the link. It can be:
#>
#> "from" means the color of the link is determined by the source node.
#> "to" means the color of the link is determined by the target node.
#> Otherwise, the color of the link is determined by the column name from the links data frame.
#>
#> - link_color_name: A character string specifying the name of the link color in the legend.
#> Only used when link_color_by is a column name.
#> - palette: A character string specifying the palette to use.
#> A named list or vector can be used to specify the palettes for different split_by values.
#> - palcolor: A character string specifying the color to use in the palette.
#> A named list can be used to specify the colors for different split_by values.
#> If some values are missing, the values from the palette will be used (palcolor will be NULL for those values).
#> - link_palette: A character string specifying the palette of the links.
#> When link_color_by is "from" or "to", the palette of the links defaults to the palette of the nodes.
#> - link_palcolor: A character vector specifying the colors of the link palette.
#> When link_color_by is "from" or "to", the colors of the link palette defaults to the colors of the node palette.
#> - directed: A logical value specifying whether the graph is directed.
#> - layout: A character string specifying the layout of the graph.
#> It can be "circle", "tree", "grid", or a layout function from igraph.
#> - cluster: A character string specifying the clustering method.
#> It can be "none", "fast_greedy", "walktrap", "edge_betweenness", "infomap", or a clustering function from igraph.
#> - add_mark: A logical value specifying whether to add mark for the clusters to the plot.
#> - mark_expand: A unit value specifying the expansion of the mark.
#> - mark_type: A character string specifying the type of the mark.
#> It can be "hull", "ellipse", "rect", "circle", or a mark function from ggforce.
#> - mark_alpha: A numeric value specifying the transparency of the mark.
#> - mark_linetype: A numeric value specifying the line type of the mark.
#> - add_label: A logical value specifying whether to add label to the nodes to the plot.
#> - label_size: A numeric value specifying the size of the label.
#> - label_fg: A character string specifying the foreground color of the label.
#> - label_bg: A character string specifying the background color of the label.
#> - label_bg_r: A numeric value specifying the background ratio of the label.
#> - arrow: An arrow object for the links.
#> - title: A character string specifying the title of the plot.
#> A function can be used to generate the title based on the default title.
#> This is useful when split_by is used and the title needs to be dynamic.
#> - subtitle: A character string specifying the subtitle of the plot.
#> - xlab: A character string specifying the x-axis label.
#> - ylab: A character string specifying the y-axis label.
#> - aspect.ratio: A numeric value specifying the aspect ratio of the plot.
#> - theme: A character string or a theme class (i.e. ggplot2::theme_classic) specifying the theme to use.
#> Default is "theme_this".
#> - theme_args: A list of arguments to pass to the theme function.
#> - legend.position: A character string specifying the position of the legend.
#> if waiver(), for single groups, the legend will be "none", otherwise "right".
#> - legend.direction: A character string specifying the direction of the legend.
#> - seed: The random seed to use. Default is 8525.
#> - combine: Whether to combine the plots into one when facet is FALSE. Default is TRUE.
#> - nrow: A numeric value specifying the number of rows in the facet.
#> - ncol: A numeric value specifying the number of columns in the facet.
#> - byrow: A logical value indicating whether to fill the plots by row.
#> - axes: A string specifying how axes should be treated. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'keep' will retain all axes in individual plots.
#> 'collect' will remove duplicated axes when placed in the same run of rows or columns of the layout.
#> 'collect_x' and 'collect_y' will remove duplicated x-axes in the columns or duplicated y-axes in the rows respectively.
#>
#> - axis_titles: A string specifying how axis titltes should be treated. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'keep' will retain all axis titles in individual plots.
#> 'collect' will remove duplicated titles in one direction and merge titles in the opposite direction.
#> 'collect_x' and 'collect_y' control this for x-axis titles and y-axis titles respectively.
#>
#> - guides: A string specifying how guides should be treated in the layout. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'collect' will collect guides below to the given nesting level, removing duplicates.
#> 'keep' will stop collection at this level and let guides be placed alongside their plot.
#> 'auto' will allow guides to be collected if a upper level tries, but place them alongside the plot if not.
#>
#> - design: Specification of the location of areas in the layout, passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE. When specified, nrow, ncol, and byrow are ignored.
#> See patchwork::wrap_plots() for more details.
#> For ChordPlot, see plotthis::ChordPlot().
#>
#> [...] can be:
#> - y: A character string specifying the column name of the data frame to plot for the y-axis.
#> - from: A character string of the column name to plot for the source.
#> A character/factor column is expected.
#> - from_sep: A character string to concatenate the columns in from, if multiple columns are provided.
#> - to: A character string of the column name to plot for the target.
#> A character/factor column is expected.
#> - to_sep: A character string to concatenate the columns in to, if multiple columns are provided.
#> - split_by_sep: The separator for multiple split_by columns. See split_by
#> - flip: A logical value to flip the source and target.
#> - links_color: A character string to specify the color of the links.
#> Either "from" or "to".
#> - theme: A character string or a theme class (i.e. ggplot2::theme_classic) specifying the theme to use.
#> Default is "theme_this".
#> - theme_args: A list of arguments to pass to the theme function.
#> - palette: A character string specifying the palette to use.
#> A named list or vector can be used to specify the palettes for different split_by values.
#> - palcolor: A character string specifying the color to use in the palette.
#> A named list can be used to specify the colors for different split_by values.
#> If some values are missing, the values from the palette will be used (palcolor will be NULL for those values).
#> - alpha: A numeric value specifying the transparency of the plot.
#> - labels_rot: A logical value to rotate the labels by 90 degrees.
#> - title: A character string specifying the title of the plot.
#> A function can be used to generate the title based on the default title.
#> This is useful when split_by is used and the title needs to be dynamic.
#> - subtitle: A character string specifying the subtitle of the plot.
#> - seed: The random seed to use. Default is 8525.
#> - combine: Whether to combine the plots into one when facet is FALSE. Default is TRUE.
#> - nrow: A numeric value specifying the number of rows in the facet.
#> - ncol: A numeric value specifying the number of columns in the facet.
#> - byrow: A logical value indicating whether to fill the plots by row.
#> - axes: A string specifying how axes should be treated. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'keep' will retain all axes in individual plots.
#> 'collect' will remove duplicated axes when placed in the same run of rows or columns of the layout.
#> 'collect_x' and 'collect_y' will remove duplicated x-axes in the columns or duplicated y-axes in the rows respectively.
#>
#> - axis_titles: A string specifying how axis titltes should be treated. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'keep' will retain all axis titles in individual plots.
#> 'collect' will remove duplicated titles in one direction and merge titles in the opposite direction.
#> 'collect_x' and 'collect_y' control this for x-axis titles and y-axis titles respectively.
#>
#> - guides: A string specifying how guides should be treated in the layout. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'collect' will collect guides below to the given nesting level, removing duplicates.
#> 'keep' will stop collection at this level and let guides be placed alongside their plot.
#> 'auto' will allow guides to be collected if a upper level tries, but place them alongside the plot if not.
#>
#> - design: Specification of the location of areas in the layout, passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE. When specified, nrow, ncol, and byrow are ignored.
#> See patchwork::wrap_plots() for more details.
#> For Heatmap, see plotthis::Heatmap().
#>
#> [...] can be:
#> - values_by: A character of column name in data that contains the values to be plotted.
#> This is required when in_form is "long". For other formats, the values are pivoted into a column named by values_by.
#> - values_fill: A value to fill in the missing values in the heatmap.
#> When there is missing value in the data, the cluster_rows and cluster_columns will fail.
#> - name: A character string to name the heatmap (will be used to rename values_by).
#> - in_form: The format of the data. Can be one of "matrix", "long", "wide-rows", "wide-columns", or "auto".
#> Defaults to "auto".
#> - split_by_sep: A character string to concat multiple columns in split_by.
#> - rows_by: A vector of column names in data that contains the row information.
#> This is used to create the rows of the heatmap.
#> When in_form is "long" or "wide-columns", this is requied, and multiple columns can be specified,
#> which will be concatenated by rows_by_sep into a single column.
#> - rows_by_sep: A character string to concat multiple columns in rows_by.
#> - rows_split_by: A character of column name in data that contains the split information for rows.
#> - rows_split_by_sep: A character string to concat multiple columns in rows_split_by.
#> - columns_by: A vector of column names in data that contains the column information.
#> This is used to create the columns of the heatmap.
#> When in_form is "long" or "wide-rows", this is required, and multiple columns can be specified,
#> which will be concatenated by columns_by_sep into a single column.
#> - columns_by_sep: A character string to concat multiple columns in columns_by.
#> - columns_split_by: A character of column name in data that contains the split information for columns.
#> - columns_split_by_sep: A character string to concat multiple columns in columns_split_by.
#> - rows_data: A data frame containing additional data for rows, which can be used to add annotations to the heatmap.
#> It will be joined to the main data by rows_by and split_by if split_by exists in rows_data.
#> This is useful for adding additional information to the rows of the heatmap.
#> - columns_data: A data frame containing additional data for columns, which can be used to add annotations to the heatmap.
#> It will be joined to the main data by columns_by and split_by if split_by exists in columns_data.
#> This is useful for adding additional information to the columns of the heatmap.
#> - columns_name: A character string to rename the column created by columns_by, which will be reflected in the name of the annotation or legend.
#> - columns_split_name: A character string to rename the column created by columns_split_by, which will be reflected in the name of the annotation or legend.
#> - rows_name: A character string to rename the column created by rows_by, which will be reflected in the name of the annotation or legend.
#> - rows_split_name: A character string to rename the column created by rows_split_by, which will be reflected in the name of the annotation or legend.
#> - palette: A character string specifying the palette of the heatmap cells.
#> - palcolor: A character vector of colors to override the palette of the heatmap cells.
#> - rows_palette: A character string specifying the palette of the row group annotation.
#> The default is "Paired".
#> - rows_palcolor: A character vector of colors to override the palette of the row group annotation.
#> - rows_split_palette: A character string specifying the palette of the row split annotation.
#> The default is "simspec".
#> - rows_split_palcolor: A character vector of colors to override the palette of the row split annotation.
#> - columns_palette: A character string specifying the palette of the column group annotation.
#> The default is "Paired".
#> - columns_palcolor: A character vector of colors to override the palette of the column group annotation.
#> - columns_split_palette: A character string specifying the palette of the column split annotation.
#> The default is "simspec".
#> - columns_split_palcolor: A character vector of colors to override the palette of the column split annotation.
#> - pie_size_name: A character string specifying the name of the legend for the pie size.
#> - pie_size: A numeric value or a function specifying the size of the pie chart.
#> If it is a function, the function should take count as the argument and return the size.
#> - pie_values: A function or character that can be converted to a function by match.arg()
#> to calculate the values for the pie chart. Default is "length".
#> The function should take a vector of values as the argument and return a single value, for each
#> group in pie_group_by.
#> - pie_name: A character string to rename the column created by pie_group_by, which will be reflected in the name of the annotation or legend.
#> - pie_group_by: A character of column name in data that contains the group information for pie charts.
#> This is used to create pie charts in the heatmap when cell_type is "pie".
#> - pie_group_by_sep: A character string to concat multiple columns in pie_group_by.
#> - pie_palette: A character string specifying the palette of the pie chart.
#> - pie_palcolor: A character vector of colors to override the palette of the pie chart.
#> - bars_sample: An integer specifying the number of samples to draw the bars.
#> - label: A function to calculate the labels for the heatmap cells.
#> It can take either 1, 3, or 5 arguments. The first argument is the aggregated values.
#> If it takes 3 arguments, the second and third arguments are the row and column indices.
#> If it takes 5 arguments, the second and third arguments are the row and column indices,
#> the fourth and fifth arguments are the row and column names.
#> The function should return a character vector of the same length as the aggregated values.
#> If the function returns NA, no label will be shown for that cell.
#> For the indices, if you have the same dimension of data (same order of rows and columns) as the heatmap, you need to use ComplexHeatmap::pindex() to get the correct values.
#> - label_size: A numeric value specifying the size of the labels when cell_type = "label".
#> - violin_fill: A character vector of colors to override the fill color of the violin plot.
#> If NULL, the fill color will be the same as the annotion.
#> - boxplot_fill: A character vector of colors to override the fill color of the boxplot.
#> If NULL, the fill color will be the same as the annotion.
#> - dot_size: A numeric value specifying the size of the dot or a function to calculate the size
#> from the values in the cell or a function to calculate the size from the values in the cell.
#> - dot_size_name: A character string specifying the name of the legend for the dot size.
#> If NULL, the dot size legend will not be shown.
#> - legend_items: A numeric vector with names to specifiy the items in the main legend.
#> The names will be working as the labels of the legend items.
#> - legend_discrete: A logical value indicating whether the main legend is discrete.
#> - legend.position: A character string specifying the position of the legend.
#> if waiver(), for single groups, the legend will be "none", otherwise "right".
#> - legend.direction: A character string specifying the direction of the legend.
#> - lower_quantile, upper_quantile, lower_cutoff, upper_cutoff: Vector of minimum and maximum cutoff values or quantile values for each feature.
#> It's applied to aggregated values when aggregated values are used (e.g. plot_type tile, label, etc).
#> It's applied to raw values when raw values are used (e.g. plot_type bars, etc).
#> - add_bg: A logical value indicating whether to add a background to the heatmap.
#> Does not work with cell_type = "bars" or cell_type = "tile".
#> - bg_alpha: A numeric value between 0 and 1 specifying the transparency of the background.
#> - add_reticle: A logical value indicating whether to add a reticle to the heatmap.
#> - reticle_color: A character string specifying the color of the reticle.
#> - column_name_annotation: A logical value indicating whether to add the column annotation for the column names.
#> which is a simple annotaion indicating the column names.
#> - column_name_legend: A logical value indicating whether to show the legend of the column name annotation.
#> - row_name_annotation: A logical value indicating whether to add the row annotation for the row names.
#> which is a simple annotaion indicating the row names.
#> - row_name_legend: A logical value indicating whether to show the legend of the row name annotation.
#> - cluster_columns: A logical value indicating whether to cluster the columns.
#> If TRUE and columns_split_by is provided, the clustering will only be applied to the columns within the same split.
#> - cluster_rows: A logical value indicating whether to cluster the rows.
#> If TRUE and rows_split_by is provided, the clustering will only be applied to the rows within the same split.
#> - border: A logical value indicating whether to draw the border of the heatmap.
#> If TRUE, the borders of the slices will be also drawn.
#> - title: The global (column) title of the heatmap
#> - column_title: A character string/vector of the column name(s) to use as the title of the column group annotation.
#> - row_title: A character string/vector of the column name(s) to use as the title of the row group annotation.
#> - na_col: A character string specifying the color for missing values.
#> The default is "grey85".
#> - row_names_side: A character string specifying the side of the row names.
#> The default is "right".
#> - column_names_side: A character string specifying the side of the column names.
#> The default is "bottom".
#> - column_annotation: A character string/vector of the column name(s) to use as the column annotation.
#> Or a list with the keys as the names of the annotation and the values as the column names.
#> - column_annotation_side: A character string specifying the side of the column annotation.
#> Could be a list with the keys as the names of the annotation and the values as the sides.
#> - column_annotation_palette: A character string specifying the palette of the column annotation.
#> The default is "Paired".
#> Could be a list with the keys as the names of the annotation and the values as the palettes.
#> - column_annotation_palcolor: A character vector of colors to override the palette of the column annotation.
#> Could be a list with the keys as the names of the annotation and the values as the palcolors.
#> - column_annotation_type: A character string specifying the type of the column annotation.
#> The default is "auto". Other options are "simple", "pie", "ring", "bar", "violin", "boxplot", "density".
#> Could be a list with the keys as the names of the annotation and the values as the types.
#> If the type is "auto", the type will be determined by the type and number of the column data.
#> - column_annotation_params: A list of parameters passed to the annotation function.
#> Could be a list with the keys as the names of the annotation and the values as the parameters passed to the annotation function. For the parameters for names (columns_by, rows_by, columns_split_by, rows_split_by), the key should be "name.(name)", where (name) is the name of the annotation.
#> See anno_pie(), anno_ring(), anno_bar(), anno_violin(), anno_boxplot(), anno_density(), anno_simple(), anno_points() and anno_lines() for the parameters of each annotation function.
#> - column_annotation_agg: A function to aggregate the values in the column annotation.
#> - row_annotation: A character string/vector of the column name(s) to use as the row annotation.
#> Or a list with the keys as the names of the annotation and the values as the column names.
#> - row_annotation_side: A character string specifying the side of the row annotation.
#> Could be a list with the keys as the names of the annotation and the values as the sides.
#> - row_annotation_palette: A character string specifying the palette of the row annotation.
#> The default is "Paired".
#> Could be a list with the keys as the names of the annotation and the values as the palettes.
#> - row_annotation_palcolor: A character vector of colors to override the palette of the row annotation.
#> Could be a list with the keys as the names of the annotation and the values as the palcolors.
#> - row_annotation_type: A character string specifying the type of the row annotation.
#> The default is "auto". Other options are "simple", "pie", "ring", "bar", "violin", "boxplot", "density".
#> Could be a list with the keys as the names of the annotation and the values as the types.
#> If the type is "auto", the type will be determined by the type and number of the row data.
#> - row_annotation_params: A list of parameters passed to the annotation function.
#> Could be a list with the keys as the names of the annotation and the values as the parameters.
#> Same as column_annotation_params.
#> - row_annotation_agg: A function to aggregate the values in the row annotation.
#> - flip: A logical value indicating whether to flip the heatmap.
#> The idea is that, you can simply set flip = TRUE to flip the heatmap.
#> You don't need to swap the arguments related to rows and columns, except those you specify via ...
#> that are passed to ComplexHeatmap::Heatmap() directly.
#> - alpha: A numeric value between 0 and 1 specifying the transparency of the heatmap cells.
#> - seed: The random seed to use. Default is 8525.
#> - layer_fun_callback: A function to add additional layers to the heatmap.
#> The function should have the following arguments: j, i, x, y, w, h, fill, sr and sc.
#> Please also refer to the layer_fun argument in ComplexHeatmap::Heatmap.
#> - cell_type: A character string specifying the type of the heatmap cells.
#> The default is values. Other options are "bars", "label", "dot", "violin", "boxplot".
#> Note that for pie chart, the values under columns specified by rows will not be used directly. Instead, the values
#> will just be counted in different pie_group_by groups. NA values will not be counted.
#> - cell_agg: A function to aggregate the values in the cell, for the cell type "tile" and "label".
#> The default is mean.
#> - combine: Whether to combine the plots into one when facet is FALSE. Default is TRUE.
#> - nrow: A numeric value specifying the number of rows in the facet.
#> - ncol: A numeric value specifying the number of columns in the facet.
#> - byrow: A logical value indicating whether to fill the plots by row.
#> - axes: A string specifying how axes should be treated. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'keep' will retain all axes in individual plots.
#> 'collect' will remove duplicated axes when placed in the same run of rows or columns of the layout.
#> 'collect_x' and 'collect_y' will remove duplicated x-axes in the columns or duplicated y-axes in the rows respectively.
#>
#> - axis_titles: A string specifying how axis titltes should be treated. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'keep' will retain all axis titles in individual plots.
#> 'collect' will remove duplicated titles in one direction and merge titles in the opposite direction.
#> 'collect_x' and 'collect_y' control this for x-axis titles and y-axis titles respectively.
#>
#> - guides: A string specifying how guides should be treated in the layout. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'collect' will collect guides below to the given nesting level, removing duplicates.
#> 'keep' will stop collection at this level and let guides be placed alongside their plot.
#> 'auto' will allow guides to be collected if a upper level tries, but place them alongside the plot if not.
#>
#> - design: Specification of the location of areas in the layout, passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE. When specified, nrow, ncol, and byrow are ignored.
#> See patchwork::wrap_plots() for more details.
#> For SankeyPlot, see plotthis::SankeyPlot().
#>
#> [...] can be:
#> - in_form: A character string to specify the format of the data.
#> Possible values are "auto", "long", "lodes", "wide", "alluvia", and "counts".
#> - x: A character string specifying the column name of the data frame to plot for the x-axis.
#> - x_sep: A character string to concatenate the columns in x, if multiple columns are provided.
#> - y: A character string specifying the column name of the data frame to plot for the y-axis.
#> - stratum: A character string of the column name to group the nodes for each x.
#> See data for more details.
#> - stratum_sep: A character string to concatenate the columns in stratum, if multiple columns are provided.
#> - alluvium: A character string of the column name to define the links.
#> See data for more details.
#> - alluvium_sep: A character string to concatenate the columns in alluvium, if multiple columns are provided.
#> - split_by_sep: The separator for multiple split_by columns. See split_by
#> - keep_empty: A logical value indicating whether to keep empty groups.
#> If FALSE, empty groups will be removed.
#> - flow: A logical value to use ggalluvial::geom_flow instead of ggalluvial::geom_alluvium.
#> - expand: The values to expand the x and y axes. It is like CSS padding.
#> When a single value is provided, it is used for both axes on both sides.
#> When two values are provided, the first value is used for the top/bottom side and the second value is used for the left/right side.
#> When three values are provided, the first value is used for the top side, the second value is used for the left/right side, and the third value is used for the bottom side.
#> When four values are provided, the values are used for the top, right, bottom, and left sides, respectively.
#> You can also use a named vector to specify the values for each side.
#> When the axis is discrete, the values will be applied as 'add' to the 'expansion' function.
#> When the axis is continuous, the values will be applied as 'mult' to the 'expansion' function.
#> See also https://ggplot2.tidyverse.org/reference/expansion.html
#> - nodes_legend: Controls how the legend of nodes will be shown. Possible values are:
#>
#> "merge": Merge the legends of nodes. That is only one legend will be shown for all nodes.
#> "separate": Show the legends of nodes separately. That is, nodes on each x will have their own legend.
#> "none": Do not show the legend of nodes.
#> "auto": Automatically determine how to show the legend.
#> When nodes_label is TRUE, "none" will apply.
#> When nodes_label is FALSE, and if stratum is the same as links_fill_by, "none" will apply.
#> If there is any overlapping values between the nodes on different x,
#> "merge" will apply. Otherwise, "separate" will apply.
#>
#> - nodes_color: A character string to color the nodes.
#> Use a special value ".fill" to use the same color as the fill.
#> - links_fill_by: A character string of the column name to fill the links.
#> - links_fill_by_sep: A character string to concatenate the columns in links_fill_by, if multiple columns are provided.
#> - links_name: A character string to name the legend of links.
#> - links_color: A character string to color the borders of links.
#> Use a special value ".fill" to use the same color as the fill.
#> - nodes_palette: A character string to specify the palette of nodes fill.
#> - nodes_palcolor: A character vector to specify the colors of nodes fill.
#> - nodes_alpha: A numeric value to specify the transparency of nodes fill.
#> - nodes_label: A logical value to show the labels on the nodes.
#> - nodes_label_miny: A numeric value to specify the minimum y (frequency) to show the labels.
#> - nodes_width: A numeric value to specify the width of nodes.
#> - links_palette: A character string to specify the palette of links fill.
#> - links_palcolor: A character vector to specify the colors of links fill.
#> - links_alpha: A numeric value to specify the transparency of links fill.
#> - legend.box: A character string to specify the box of the legend, either "vertical" or "horizontal".
#> - aspect.ratio: A numeric value specifying the aspect ratio of the plot.
#> - legend.position: A character string specifying the position of the legend.
#> if waiver(), for single groups, the legend will be "none", otherwise "right".
#> - legend.direction: A character string specifying the direction of the legend.
#> - flip: A logical value to flip the plot.
#> - theme: A character string or a theme class (i.e. ggplot2::theme_classic) specifying the theme to use.
#> Default is "theme_this".
#> - theme_args: A list of arguments to pass to the theme function.
#> - title: A character string specifying the title of the plot.
#> A function can be used to generate the title based on the default title.
#> This is useful when split_by is used and the title needs to be dynamic.
#> - subtitle: A character string specifying the subtitle of the plot.
#> - xlab: A character string specifying the x-axis label.
#> - ylab: A character string specifying the y-axis label.
#> - facet_scales: Whether to scale the axes of facets. Default is "fixed"
#> Other options are "free", "free_x", "free_y". See ggplot2::facet_wrap
#> - facet_ncol: A numeric value specifying the number of columns in the facet.
#> When facet_by is a single column and facet_wrap is used.
#> - facet_nrow: A numeric value specifying the number of rows in the facet.
#> When facet_by is a single column and facet_wrap is used.
#> - facet_byrow: A logical value indicating whether to fill the plots by row. Default is TRUE.
#> - seed: The random seed to use. Default is 8525.
#> - combine: Whether to combine the plots into one when facet is FALSE. Default is TRUE.
#> - nrow: A numeric value specifying the number of rows in the facet.
#> - ncol: A numeric value specifying the number of columns in the facet.
#> - byrow: A logical value indicating whether to fill the plots by row.
#> - axes: A string specifying how axes should be treated. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'keep' will retain all axes in individual plots.
#> 'collect' will remove duplicated axes when placed in the same run of rows or columns of the layout.
#> 'collect_x' and 'collect_y' will remove duplicated x-axes in the columns or duplicated y-axes in the rows respectively.
#>
#> - axis_titles: A string specifying how axis titltes should be treated. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'keep' will retain all axis titles in individual plots.
#> 'collect' will remove duplicated titles in one direction and merge titles in the opposite direction.
#> 'collect_x' and 'collect_y' control this for x-axis titles and y-axis titles respectively.
#>
#> - guides: A string specifying how guides should be treated in the layout. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'collect' will collect guides below to the given nesting level, removing duplicates.
#> 'keep' will stop collection at this level and let guides be placed alongside their plot.
#> 'auto' will allow guides to be collected if a upper level tries, but place them alongside the plot if not.
#>
#> - design: Specification of the location of areas in the layout, passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE. When specified, nrow, ncol, and byrow are ignored.
#> See patchwork::wrap_plots() for more details.
#> For DotPlot, see plotthis::DotPlot().
#>
#> [...] can be:
#> - x: A character vector specifying the column to use for the x-axis.
#> A numeric column is expected.
#> - y: A character vector specifying the column to use for the y-axis.
#> A factor/character column is expected.
#> - x_sep: A character vector to concatenate multiple columns in x. Default is "_".
#> - y_sep: A character vector to concatenate multiple columns in y. Default is "_".
#> - flip: A logical value indicating whether to flip the x and y axes. Default is FALSE.
#> - split_by_sep: The separator for multiple split_by columns. See split_by
#> - size_name: A character vector specifying the name for the size legend.
#> - fill_name: A character vector specifying the name for the fill legend.
#> - fill_cutoff_name: A character vector specifying the name for the fill cutoff legend.
#> - add_bg: A logical value indicating whether to add a background color to the plot. Default is FALSE.
#> - bg_palette: A character vector specifying the palette for the background color. Default is "stripe".
#> - bg_palcolor: A character vector specifying the color for the background color.
#> - bg_alpha: A numeric value specifying the alpha for the background color. Default is 0.2.
#> - bg_direction: A character vector specifying the direction for the background color. Default is "vertical".
#> Other options are "horizontal". "h" and "v" are also accepted.
#> - size_by: Which column to use as the size of the dots. It must be a numeric column.
#> If not provided, the size will be the count of the instances for each 'y' in 'x'.
#> For 'ScatterPlot', it can be a single numeric value to specify the size of the dots.
#> - fill_by: Which column to use as the fill the dots. It must be a numeric column.
#> If not provided, all dots will be filled with the same color at the middle of the palette.
#> - fill_cutoff: A numeric value specifying the cutoff for the fill column.
#> - fill_reverse: A logical value indicating whether to reverse the fill direction. Default is FALSE.
#> By default, the fill direction is "up". If TRUE, the fill direction is "down".
#> When the direction is "up", the values less than the cutoff will be filled with grey.
#> When the direction is "down", the values greater than the cutoff will be filled with grey.
#> - theme: A character string or a theme class (i.e. ggplot2::theme_classic) specifying the theme to use.
#> Default is "theme_this".
#> - theme_args: A list of arguments to pass to the theme function.
#> - palette: A character string specifying the palette to use.
#> A named list or vector can be used to specify the palettes for different split_by values.
#> - palcolor: A character string specifying the color to use in the palette.
#> A named list can be used to specify the colors for different split_by values.
#> If some values are missing, the values from the palette will be used (palcolor will be NULL for those values).
#> - alpha: A numeric value specifying the transparency of the plot.
#> - facet_scales: Whether to scale the axes of facets. Default is "fixed"
#> Other options are "free", "free_x", "free_y". See ggplot2::facet_wrap
#> - facet_ncol: A numeric value specifying the number of columns in the facet.
#> When facet_by is a single column and facet_wrap is used.
#> - facet_nrow: A numeric value specifying the number of rows in the facet.
#> When facet_by is a single column and facet_wrap is used.
#> - facet_byrow: A logical value indicating whether to fill the plots by row. Default is TRUE.
#> - seed: The random seed to use. Default is 8525.
#> - aspect.ratio: A numeric value specifying the aspect ratio of the plot.
#> - legend.position: A character string specifying the position of the legend.
#> if waiver(), for single groups, the legend will be "none", otherwise "right".
#> - legend.direction: A character string specifying the direction of the legend.
#> - title: A character string specifying the title of the plot.
#> A function can be used to generate the title based on the default title.
#> This is useful when split_by is used and the title needs to be dynamic.
#> - subtitle: A character string specifying the subtitle of the plot.
#> - xlab: A character string specifying the x-axis label.
#> - ylab: A character string specifying the y-axis label.
#> - keep_empty: A logical value indicating whether to keep empty groups.
#> If FALSE, empty groups will be removed.
#> - combine: Whether to combine the plots into one when facet is FALSE. Default is TRUE.
#> - nrow: A numeric value specifying the number of rows in the facet.
#> - ncol: A numeric value specifying the number of columns in the facet.
#> - byrow: A logical value indicating whether to fill the plots by row.
#> - axes: A string specifying how axes should be treated. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'keep' will retain all axes in individual plots.
#> 'collect' will remove duplicated axes when placed in the same run of rows or columns of the layout.
#> 'collect_x' and 'collect_y' will remove duplicated x-axes in the columns or duplicated y-axes in the rows respectively.
#>
#> - axis_titles: A string specifying how axis titltes should be treated. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'keep' will retain all axis titles in individual plots.
#> 'collect' will remove duplicated titles in one direction and merge titles in the opposite direction.
#> 'collect_x' and 'collect_y' control this for x-axis titles and y-axis titles respectively.
#>
#> - guides: A string specifying how guides should be treated in the layout. Passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE.
#> Options are:
#>
#> 'collect' will collect guides below to the given nesting level, removing duplicates.
#> 'keep' will stop collection at this level and let guides be placed alongside their plot.
#> 'auto' will allow guides to be collected if a upper level tries, but place them alongside the plot if not.
#>
#> - design: Specification of the location of areas in the layout, passed to patchwork::wrap_plots().
#> Only relevant when split_by is used and combine is TRUE. When specified, nrow, ncol, and byrow are ignored.
#> See patchwork::wrap_plots() for more details.
#>
#> - value
#>
#> A ggplot object or a list if combine is FALSE
#>
#> - examples
#>
#>
#> set.seed(8525)
#> data(cellphonedb_res)
#> CCCPlot(data = cellphonedb_res, plot_type = "network", legend.position = "none",
#> theme = "theme_blank", theme_args = list(add_coord = FALSE))
#> CCCPlot(cellphonedb_res, plot_type = "chord")
#> CCCPlot(cellphonedb_res, plot_type = "heatmap")
#> CCCPlot(cellphonedb_res, plot_type = "dot",
#> magnitude_agg = mean, magnitude_name = "Average Interaction Strength")
#> CCCPlot(cellphonedb_res, plot_type = "sankey")
#>
#> cellphonedb_res_sub <- cellphonedb_res[
#> cellphonedb_res$source %in% c("Dendritic", "CD14+ Monocyte"),]
#> CCCPlot(cellphonedb_res_sub, plot_type = "dot", method = "interaction")
#> CCCPlot(cellphonedb_res_sub, plot_type = "network", method = "interaction",
#> node_size_by = 1)
#> CCCPlot(cellphonedb_res_sub, plot_type = "heatmap", method = "interaction",
#> palette = "Reds")
#> CCCPlot(cellphonedb_res_sub, plot_type = "box", method = "interaction")
#> CCCPlot(cellphonedb_res_sub, plot_type = "violin", method = "interaction",
#> add_box = TRUE)
#> CCCPlot(cellphonedb_res_sub, plot_type = "ridge", method = "interaction")
#>
#>
#>
#>
#> You must code in the programming language 'R' to answer this prompt.
#> You can use functions from these packages: scplotter.
#> You may not install or load any additional packages.
#> These objects already exist in the R session:
#>
#> Object_name, Type
#> cellphonedb_res, data.frame.
#>
#> Do not define these objects in your R code.#> --- Receiving response from LLM provider: ---#> ```r
#> CCCPlot(data = cellphonedb_res, plot_type = "network")
#> ```#> Code ran:
#> CCCPlot(data = cellphonedb_res, plot_type = "network")