Plot the residency of the clones in different samples.
Usage
ClonalResidencyPlot(
data,
clone_call = "aa",
chain = "both",
plot_type = c("scatter", "venn", "upset"),
group_by = "Sample",
groups = NULL,
facet_by = NULL,
split_by = NULL,
split_by_sep = "_",
scatter_cor = "pearson",
scatter_size_by = c("max", "total"),
order = list(),
combine = TRUE,
nrow = NULL,
ncol = NULL,
byrow = TRUE,
...
)Arguments
- data
The product of scRepertoire::combineTCR, scRepertoire::combineTCR, or scRepertoire::combineExpression.
- clone_call
How to call the clone - VDJC gene (gene), CDR3 nucleotide (nt), CDR3 amino acid (aa), VDJC gene + CDR3 nucleotide (strict) or a custom variable in the data
- chain
indicate if both or a specific chain should be used - e.g. "both", "TRA", "TRG", "IGH", "IGL"
- plot_type
The type of plot to use. Default is "scatter". Possible values are "scatter", "venn", and "upset".
- group_by
The column name in the meta data to group the cells. Default: "Sample"
- groups
The groups to compare. Default is NULL. If NULL, all the groups in
group_bywill be compared. Note that for "scatter" plot, only two groups can be compared. So when there are more than two groups, the combination of the pairs will be used. For "scatter" plot, the groups can be specified as the comparisons separated by ":", e.g. "L:B", "Y:X".- facet_by
The column name in the meta data to facet the plots. Default: NULL
- split_by
The column name in the meta data to split the plots. Default: NULL
- split_by_sep
The separator used to concatenate the split_by when multiple columns are used.
- scatter_cor
The correlation method to use for the scatter plot. Default is "pearson".
- scatter_size_by
The size of the points in the scatter plot. Default is "max". Possible values are "max" and "total".
"max" - The max size of the clone in the two groups.
"total" - The total size of the clone in the two groups.
- order
The order of the x-axis items or groups. Default is an empty list. It should be a list of values. The names are the column names, and the values are the order.
- combine
Whether to combine the plots into a single plot. Default is TRUE.
- nrow
The number of rows in the combined plot. Default is NULL.
- ncol
The number of columns in the combined plot. Default is NULL.
- byrow
Whether to fill the combined plot by row. Default is TRUE.
- ...
Other arguments passed to the specific plot function.
For
scatterplot, seeplotthis::ScatterPlot().For
vennplot, seeplotthis::VennDiagram().For
upsetplot, seeplotthis::UpsetPlot().
Examples
# \donttest{
set.seed(8525)
data(contig_list, package = "scRepertoire")
data <- scRepertoire::combineTCR(contig_list,
samples = c("P17B", "P17L", "P18B", "P18L", "P19B","P19L", "P20B", "P20L"))
data <- scRepertoire::addVariable(data,
variable.name = "Type",
variables = rep(c("B", "L", "X", "Y"), 2)
)
data <- scRepertoire::addVariable(data,
variable.name = "Subject",
variables = rep(c("P17", "P18", "P19", "P20"), each = 2)
)
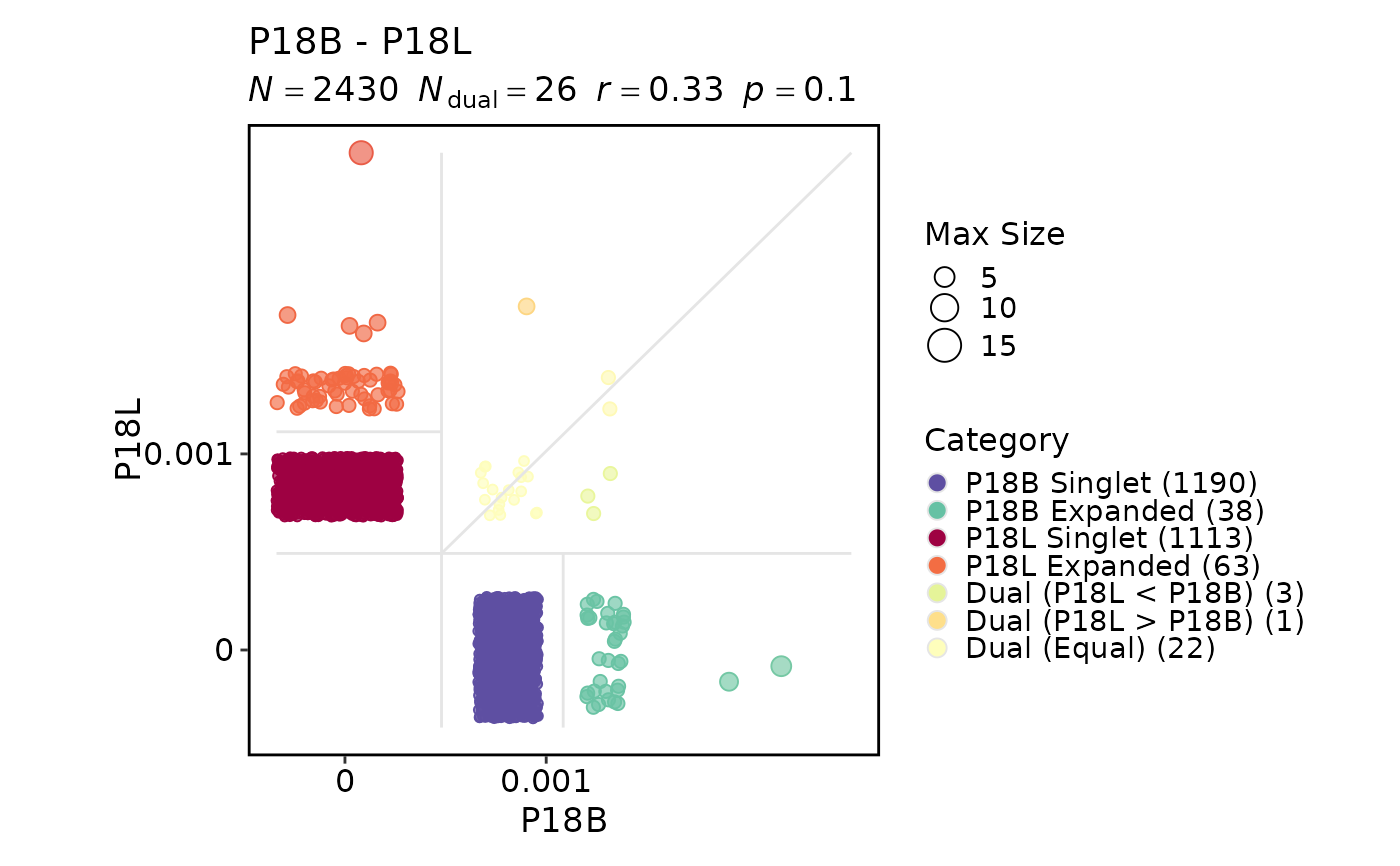
ClonalResidencyPlot(data, groups = c("P18B", "P18L"))
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_point()`).
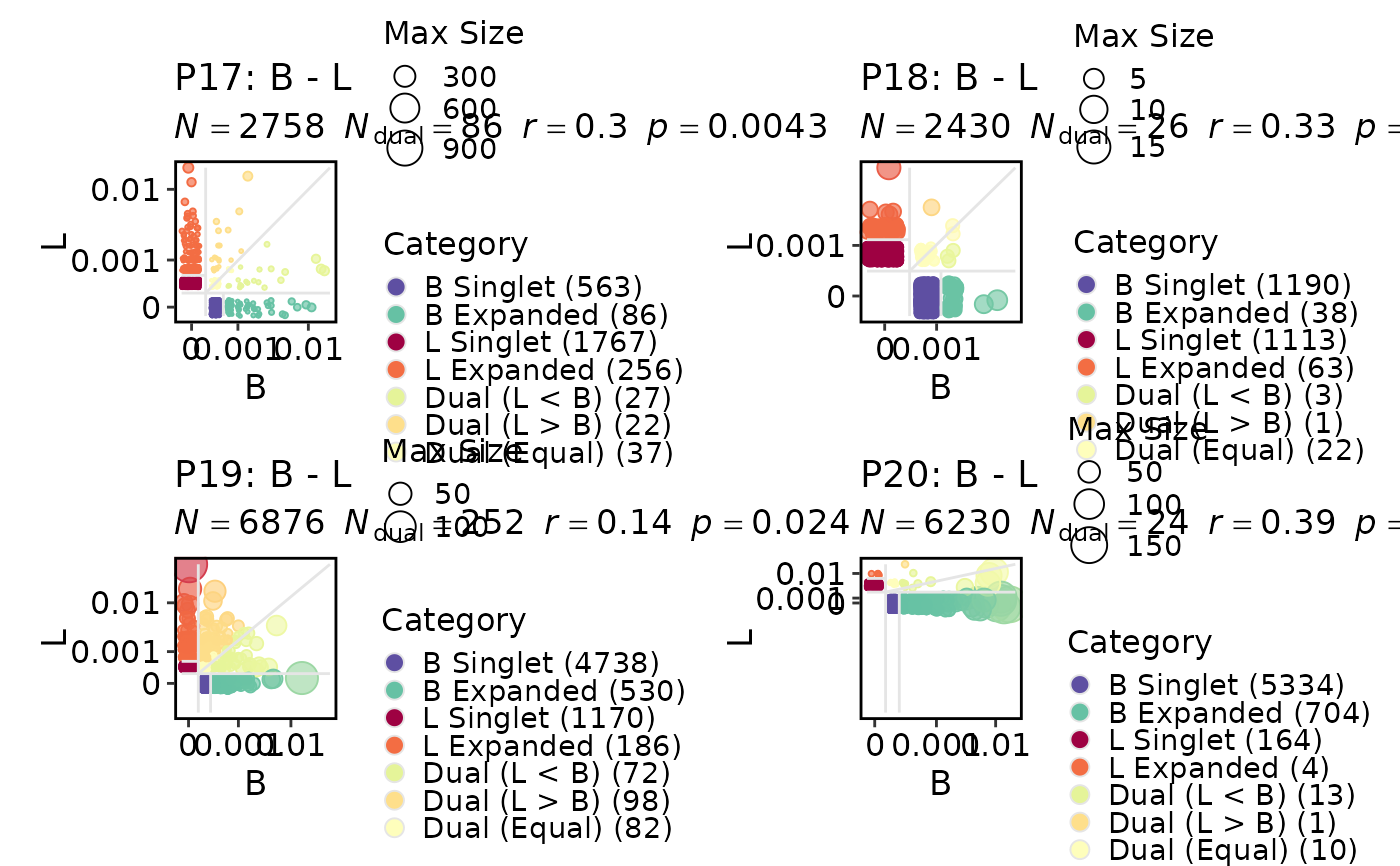
 ClonalResidencyPlot(data, group_by = "Type", groups = c("L", "B"),
split_by = "Subject")
#> Warning: [ClonalResidencyPlot] Not both groups 'L, B' is not present in the data for 'P18'. Skipping.
#> Warning: [ClonalResidencyPlot] Not both groups 'L, B' is not present in the data for 'P20'. Skipping.
#> Warning: Removed 8 rows containing missing values or values outside the scale range
#> (`geom_point()`).
#> Warning: Removed 3 rows containing missing values or values outside the scale range
#> (`geom_point()`).
ClonalResidencyPlot(data, group_by = "Type", groups = c("L", "B"),
split_by = "Subject")
#> Warning: [ClonalResidencyPlot] Not both groups 'L, B' is not present in the data for 'P18'. Skipping.
#> Warning: [ClonalResidencyPlot] Not both groups 'L, B' is not present in the data for 'P20'. Skipping.
#> Warning: Removed 8 rows containing missing values or values outside the scale range
#> (`geom_point()`).
#> Warning: Removed 3 rows containing missing values or values outside the scale range
#> (`geom_point()`).
 ClonalResidencyPlot(data, group_by = "Type", groups = c("L:B", "Y:X"),
split_by = "Subject")
#> Warning: [ClonalResidencyPlot] Not both groups 'Y, X' is not present in the data for 'P17'. Skipping.
#> Warning: [ClonalResidencyPlot] Not both groups 'L, B' is not present in the data for 'P18'. Skipping.
#> Warning: [ClonalResidencyPlot] Not both groups 'Y, X' is not present in the data for 'P19'. Skipping.
#> Warning: [ClonalResidencyPlot] Not both groups 'L, B' is not present in the data for 'P20'. Skipping.
#> Warning: Removed 8 rows containing missing values or values outside the scale range
#> (`geom_point()`).
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_point()`).
#> Warning: Removed 3 rows containing missing values or values outside the scale range
#> (`geom_point()`).
#> Warning: Removed 201 rows containing missing values or values outside the scale range
#> (`geom_point()`).
#> Warning: Removed 1 row containing missing values or values outside the scale range
#> (`geom_point()`).
ClonalResidencyPlot(data, group_by = "Type", groups = c("L:B", "Y:X"),
split_by = "Subject")
#> Warning: [ClonalResidencyPlot] Not both groups 'Y, X' is not present in the data for 'P17'. Skipping.
#> Warning: [ClonalResidencyPlot] Not both groups 'L, B' is not present in the data for 'P18'. Skipping.
#> Warning: [ClonalResidencyPlot] Not both groups 'Y, X' is not present in the data for 'P19'. Skipping.
#> Warning: [ClonalResidencyPlot] Not both groups 'L, B' is not present in the data for 'P20'. Skipping.
#> Warning: Removed 8 rows containing missing values or values outside the scale range
#> (`geom_point()`).
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_point()`).
#> Warning: Removed 3 rows containing missing values or values outside the scale range
#> (`geom_point()`).
#> Warning: Removed 201 rows containing missing values or values outside the scale range
#> (`geom_point()`).
#> Warning: Removed 1 row containing missing values or values outside the scale range
#> (`geom_point()`).
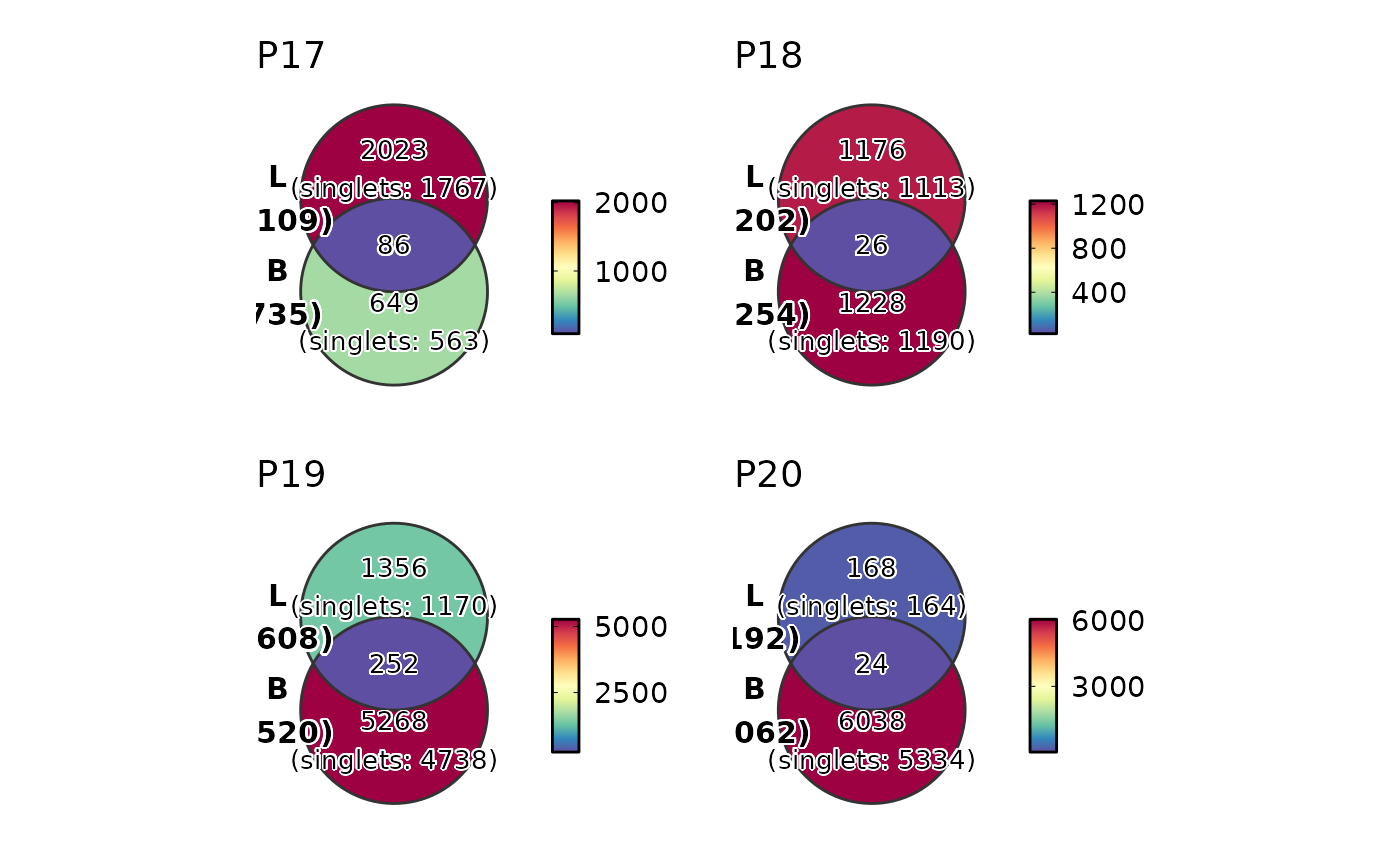 ClonalResidencyPlot(data, plot_type = "venn", groups = c("B", "L"),
group_by = "Type", split_by = "Subject")
ClonalResidencyPlot(data, plot_type = "venn", groups = c("B", "L"),
group_by = "Type", split_by = "Subject")
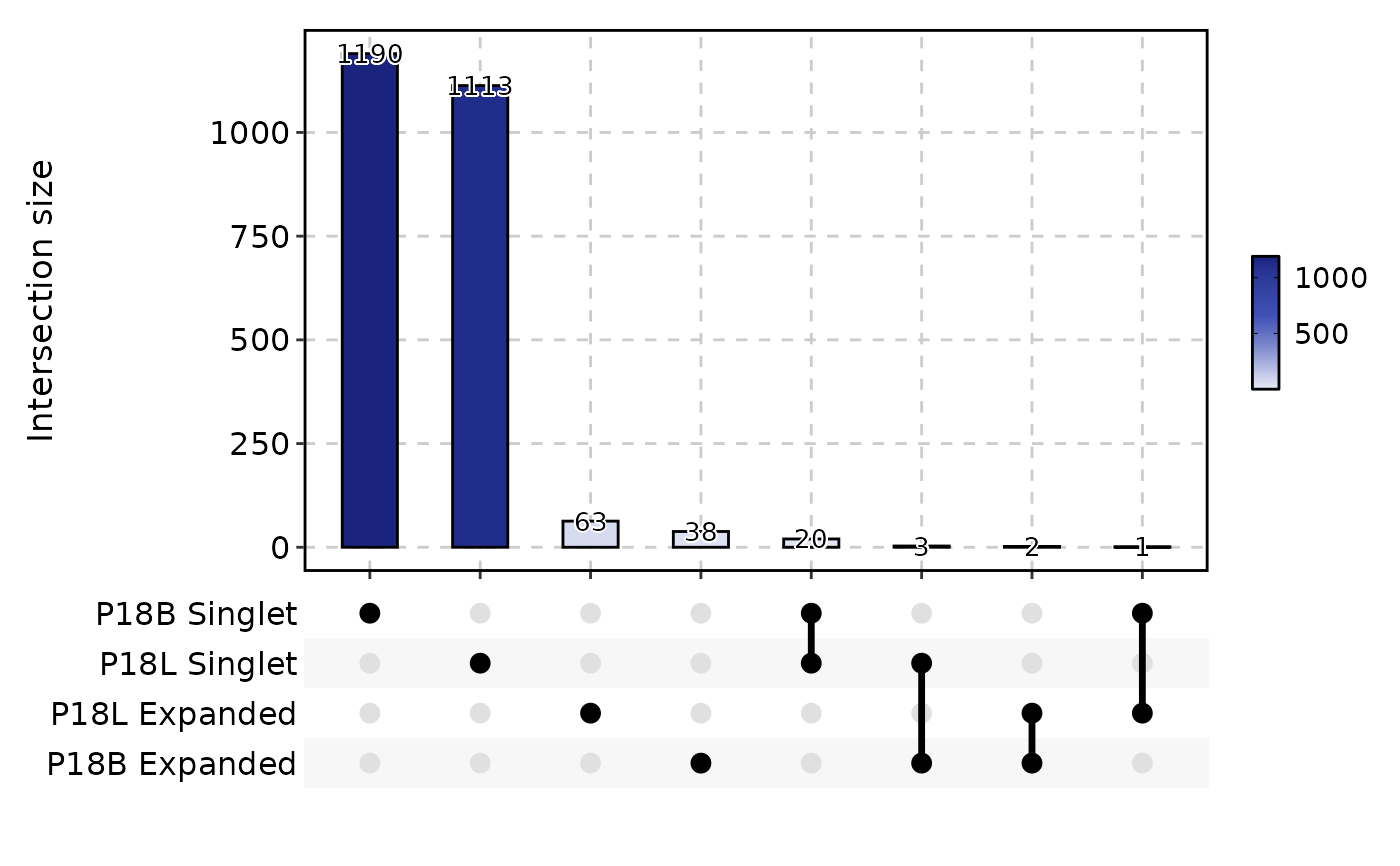 ClonalResidencyPlot(data, plot_type = "upset", groups = c("P18B", "P18L"))
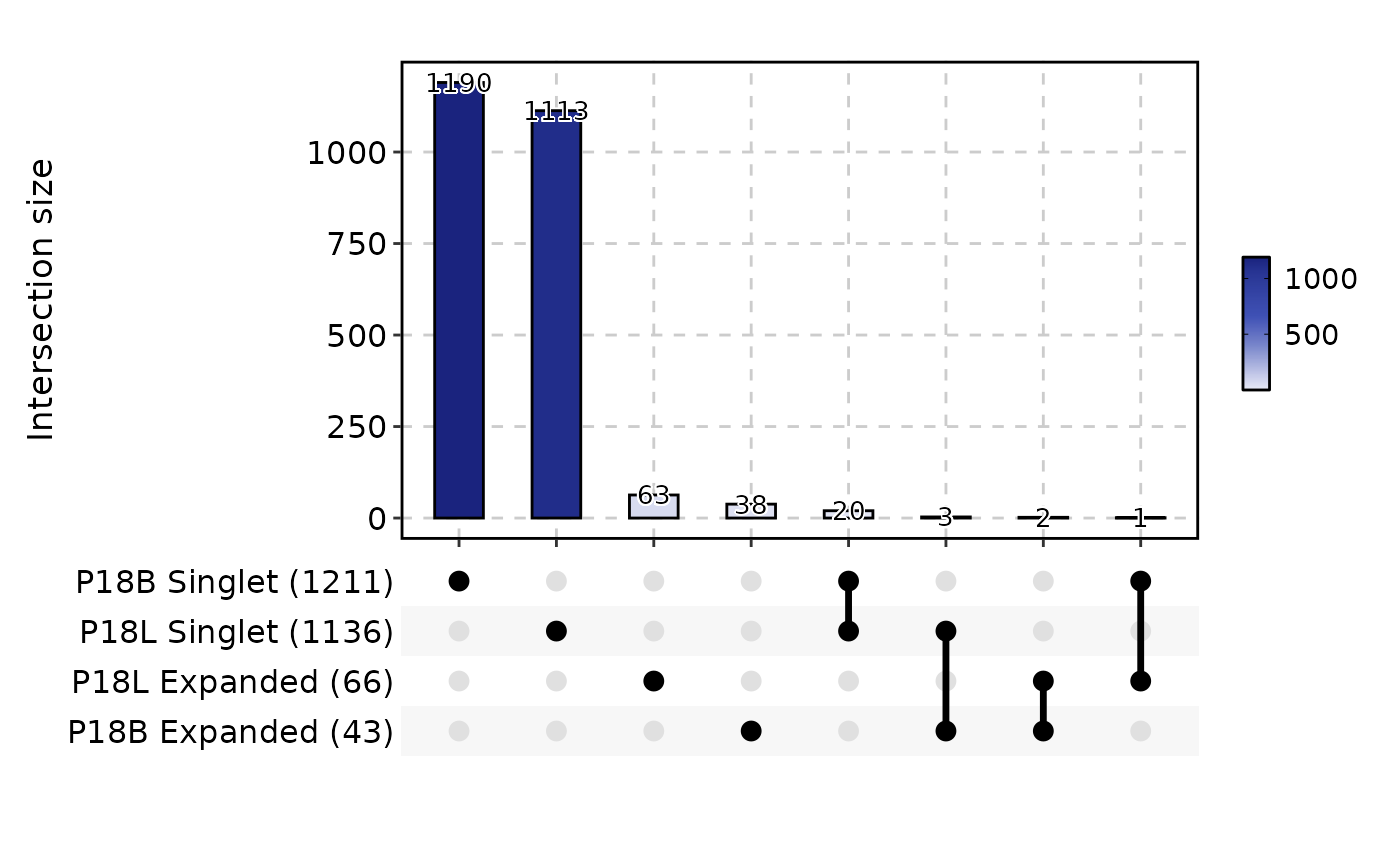
ClonalResidencyPlot(data, plot_type = "upset", groups = c("P18B", "P18L"))
 # }
# }