Plot the clonal diversities of the samples/groups.
Usage
ClonalDiversityPlot(
data,
clone_call = "gene",
chain = "both",
method = c("shannon", "gini.coeff", "inv.simpson", "norm.entropy", "gini.simpson",
"chao1", "ACE", "d50", "dXX"),
d = 50,
plot_type = c("bar", "box", "violin"),
position = "dodge",
group_by = NULL,
facet_by = NULL,
split_by = NULL,
xlab = NULL,
ylab = NULL,
...
)Arguments
- data
The product of scRepertoire::combineTCR, scRepertoire::combineTCR, or scRepertoire::combineExpression.
- clone_call
How to call the clone - VDJC gene (gene), CDR3 nucleotide (nt), CDR3 amino acid (aa), VDJC gene + CDR3 nucleotide (strict) or a custom variable in the data
- chain
indicate if both or a specific chain should be used - e.g. "both", "TRA", "TRG", "IGH", "IGL"
- method
The method to calculate the diversity. Options are "shannon" (default), "inv.simpson", "norm.entropy", "gini.simpson", "chao1", "ACE", "gini.coeff", "d50" and "dXX". See scRepertoire::clonalDiversity for details. The last 3 methods are supported by
scplotteronly:"gini.coeff" - The Gini Coefficient. A measure of inequality in the distribution of clones. 0 indicates perfect equality, 1 indicates perfect inequality.
"d50" - The number of clones that make up
50%of the total number of clones."dXX" - The number of clones that make up
XX%of the total number of clones.
- d
The percentage for the "dXX" method. Default is 50.
- plot_type
The type of plot. Options are "bar", "box" and "violin".
- position
The position adjustment for the bars. Default is "dodge".
- group_by
A character vector of column names to group the samples. Default is NULL.
- facet_by
A character vector of column names to facet the plots. Default is NULL.
- split_by
A character vector of column names to split the plots. Default is NULL.
- xlab
The x-axis label. Default is NULL.
- ylab
The y-axis label. Default is NULL.
- ...
Other arguments passed to the specific plot function.
For "bar",
plotthis::BarPlot().For "box",
plotthis::BoxPlot().For "violin",
plotthis::ViolinPlot().
Examples
set.seed(8525)
data(contig_list, package = "scRepertoire")
data <- scRepertoire::combineTCR(contig_list,
samples = c("P17B", "P17L", "P18B", "P18L", "P19B","P19L", "P20B", "P20L"))
data <- scRepertoire::addVariable(data,
variable.name = "Type",
variables = rep(c("B", "L"), 4)
)
data <- scRepertoire::addVariable(data,
variable.name = "Subject",
variables = rep(c("P17", "P18", "P19", "P20"), each = 2)
)
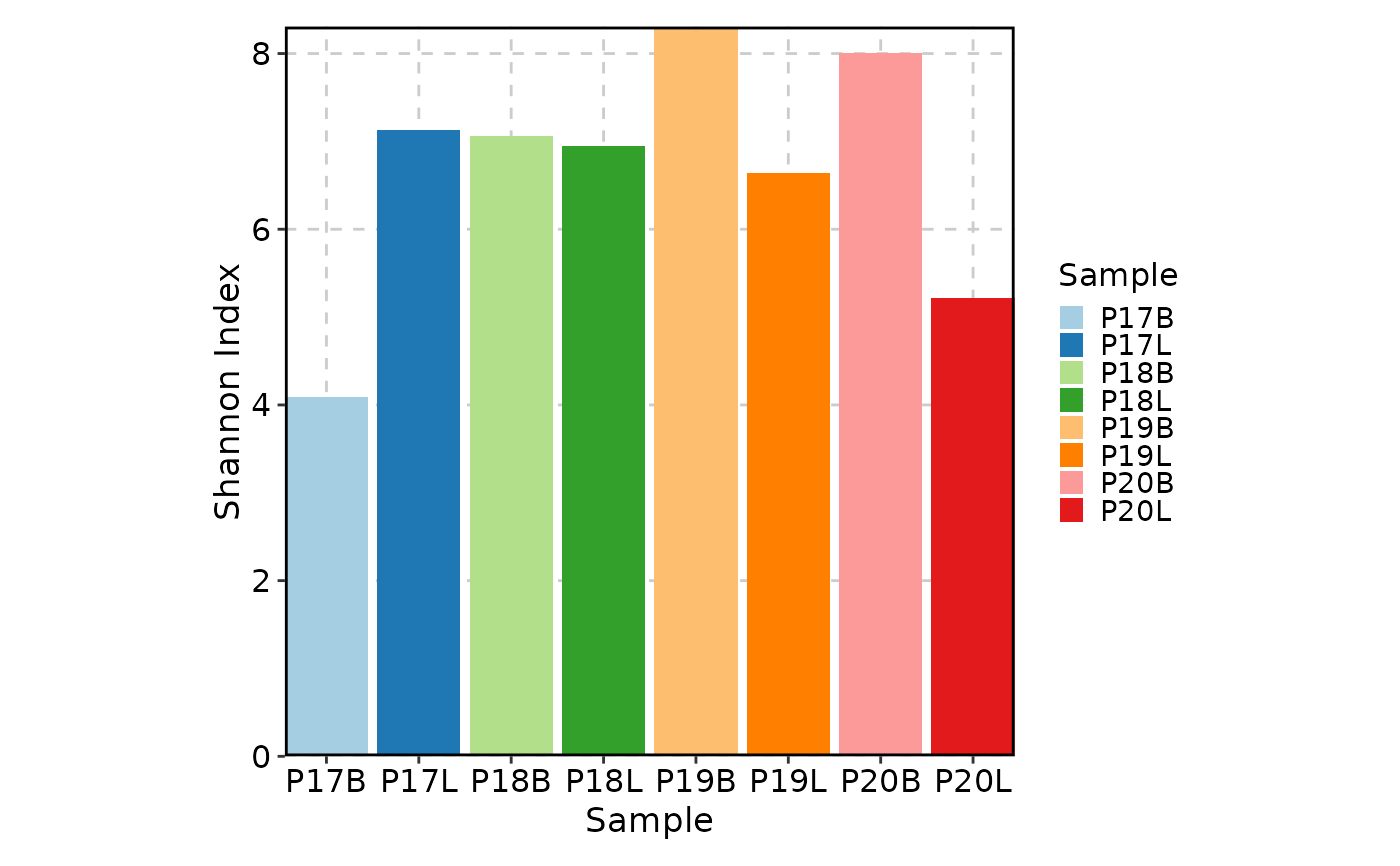
ClonalDiversityPlot(data)
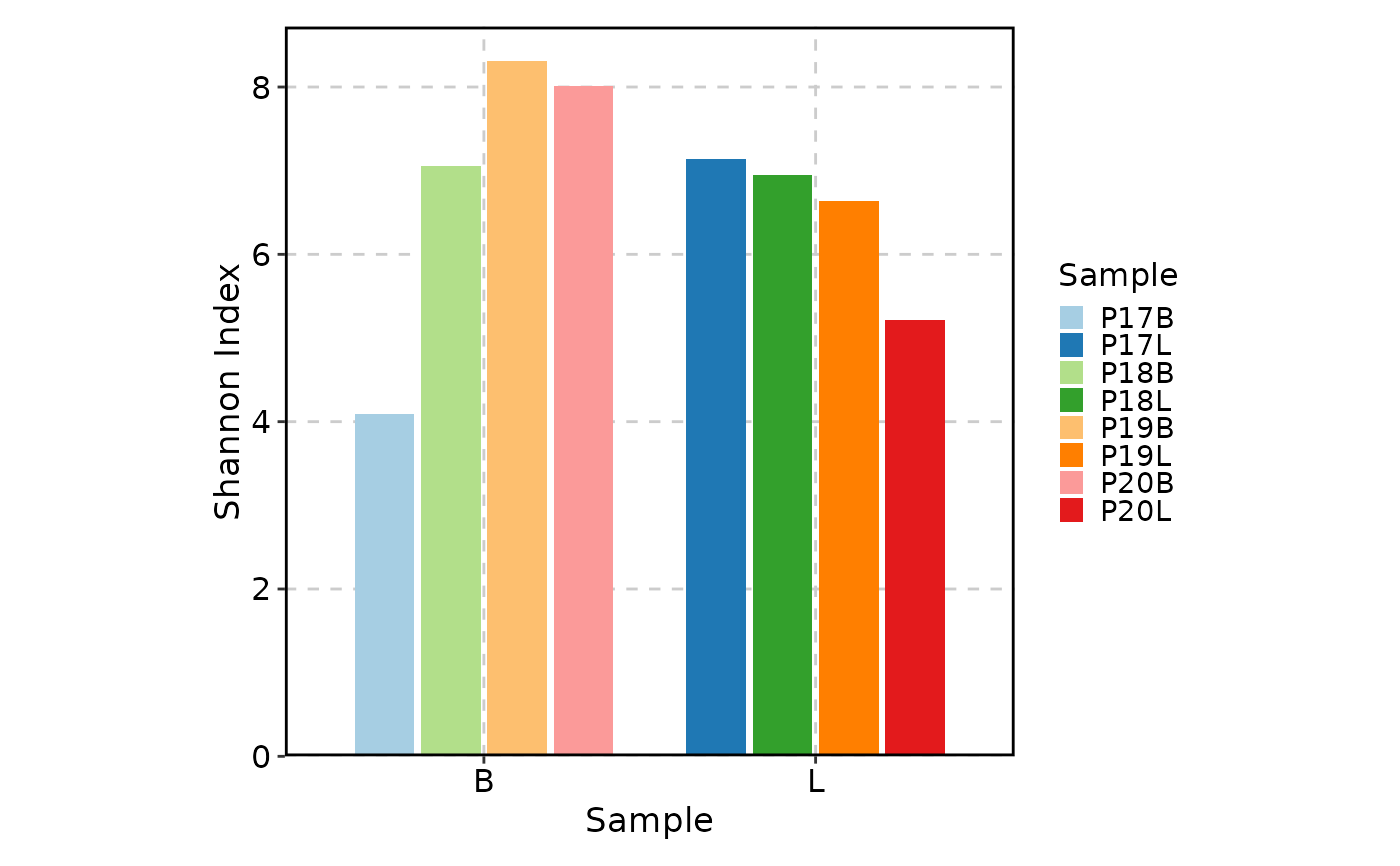
 ClonalDiversityPlot(data, group_by = "Type")
ClonalDiversityPlot(data, group_by = "Type")
 ClonalDiversityPlot(data, group_by = "Type", plot_type = "box")
ClonalDiversityPlot(data, group_by = "Type", plot_type = "box")
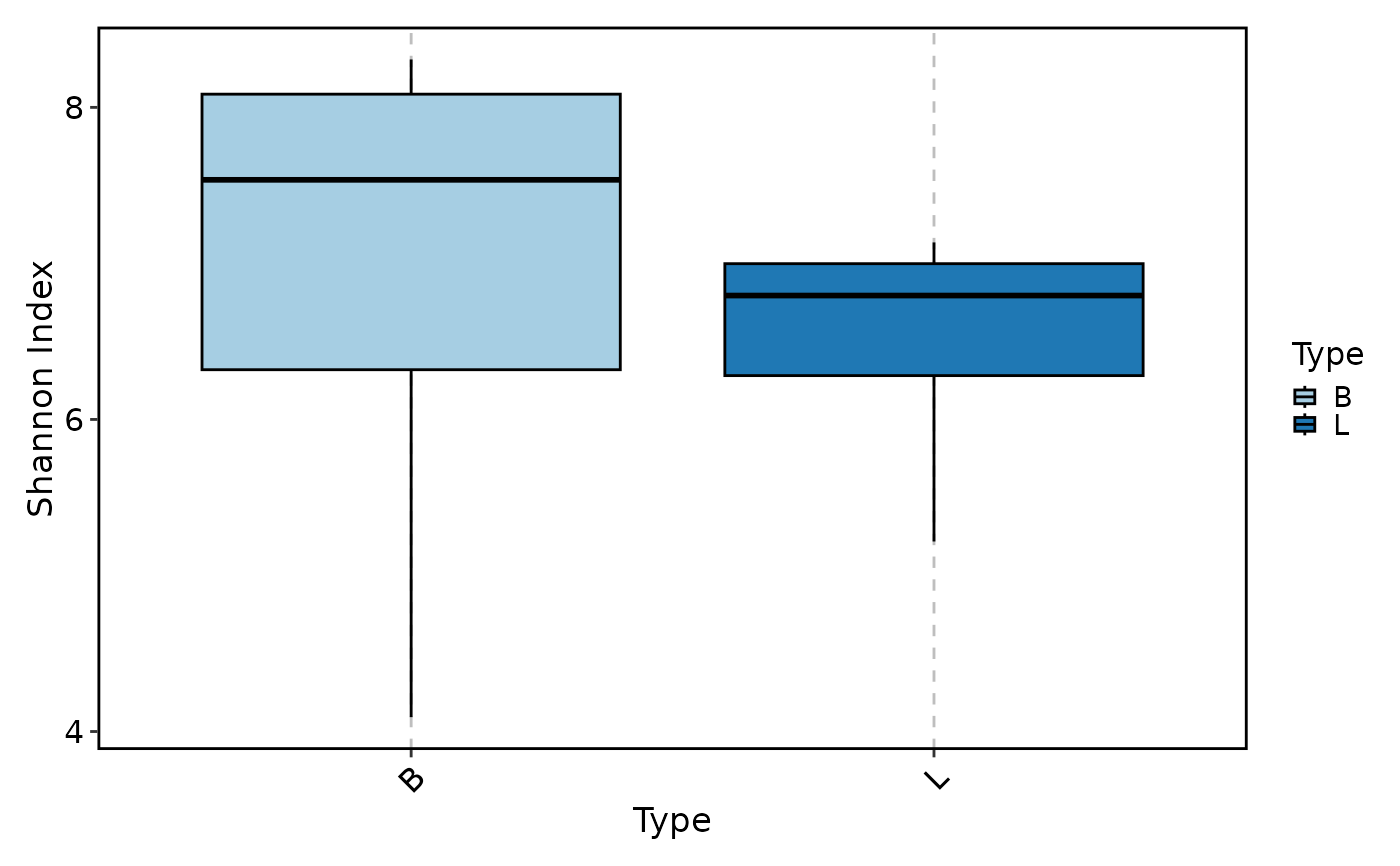 ClonalDiversityPlot(data, group_by = "Type", plot_type = "violin")
ClonalDiversityPlot(data, group_by = "Type", plot_type = "violin")
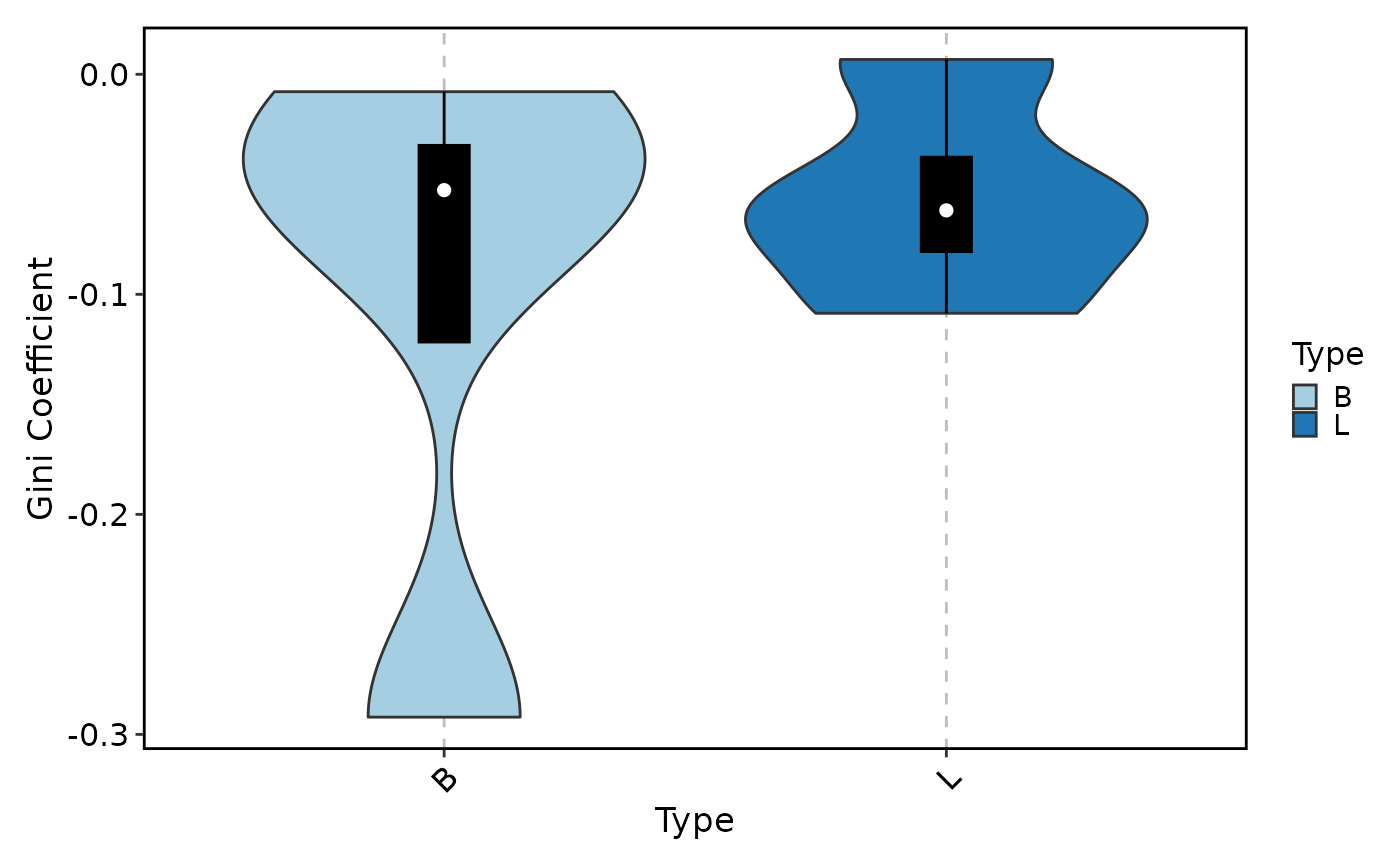
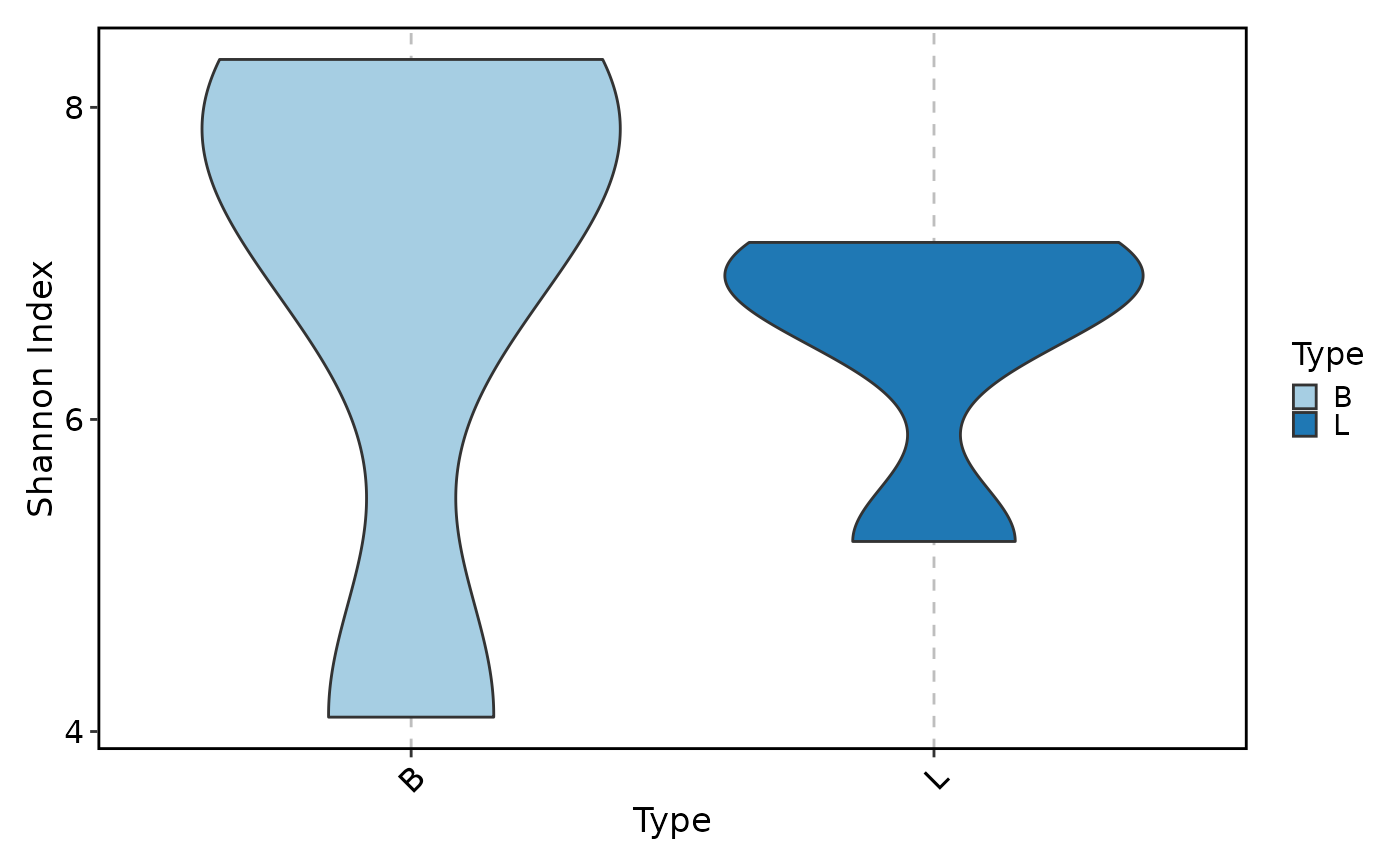 ClonalDiversityPlot(data, group_by = "Type", plot_type = "violin",
method = "gini.coeff", add_box = TRUE)
ClonalDiversityPlot(data, group_by = "Type", plot_type = "violin",
method = "gini.coeff", add_box = TRUE)
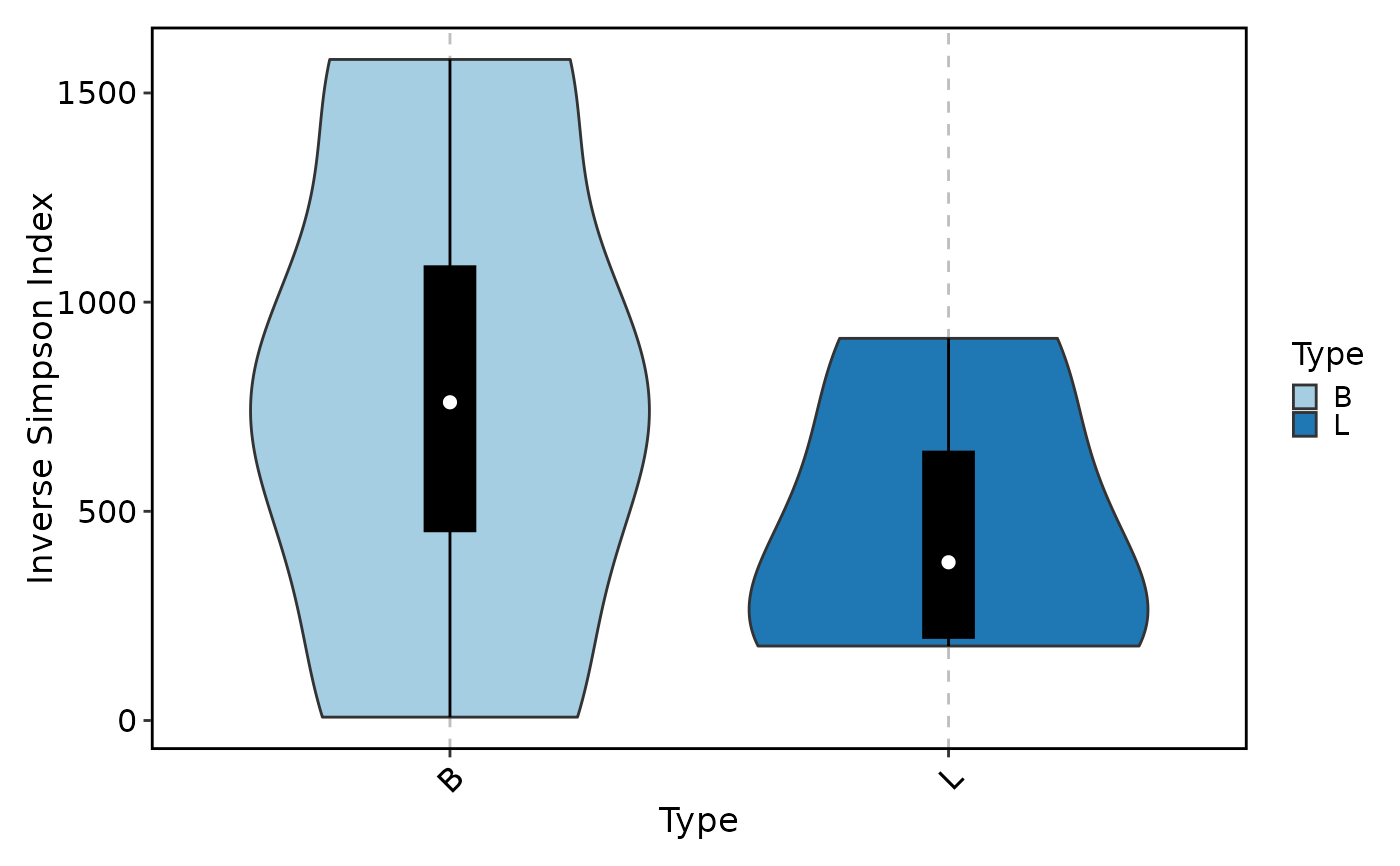
 ClonalDiversityPlot(data, group_by = "Type", plot_type = "violin",
method = "inv.simpson", add_box = TRUE)
ClonalDiversityPlot(data, group_by = "Type", plot_type = "violin",
method = "inv.simpson", add_box = TRUE)
 ClonalDiversityPlot(data, group_by = "Type", plot_type = "violin",
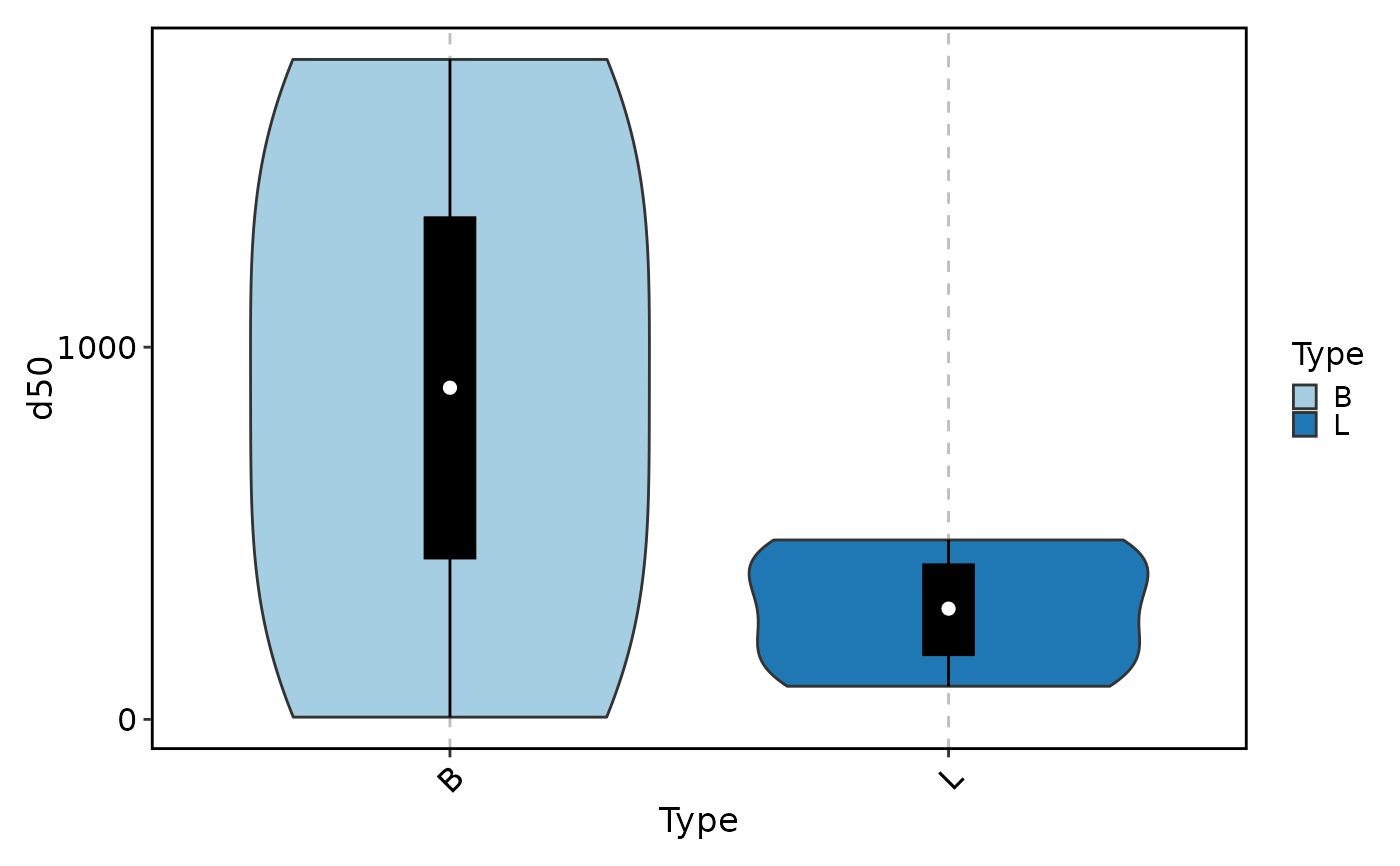
method = "d50", add_box = TRUE)
ClonalDiversityPlot(data, group_by = "Type", plot_type = "violin",
method = "d50", add_box = TRUE)