ClonalVolumePlot
Arguments
- data
The product of scRepertoire::combineTCR, scRepertoire::combineTCR, or scRepertoire::combineExpression.
- clone_call
How to call the clone - VDJC gene (gene), CDR3 nucleotide (nt), CDR3 amino acid (aa), VDJC gene + CDR3 nucleotide (strict) or a custom variable in the data
- chain
indicate if both or a specific chain should be used - e.g. "both", "TRA", "TRG", "IGH", "IGL"
- scale
Whether to use clone proportion or clone size for the plot.
- plot_type
The type of plot to use. Default is "bar". Possible values are "bar", "box", and "violin". When "box" or "violin" is used, the data will be broken down by the Sample and plotted for each group.
- x
The column name in the meta data to use as the x-axis. Default: "Sample"
- group_by
The column name in the meta data to group the cells. Default: NULL
- facet_by
The column name in the meta data to facet the plots. Default: NULL
- split_by
The column name in the meta data to split the plots. Default: NULL
- order
The order of the x-axis items or groups. Default is an empty list. It should be a list of values. The names are the column names, and the values are the order.
- ylab
The y-axis label.
- ...
Other arguments passed to the specific plot function.
For
barplot, seeplotthis::BarPlot().For
boxplot, seeplotthis::BoxPlot().For
violinplot, seeplotthis::ViolinPlot().
Examples
# \donttest{
set.seed(8525)
data(contig_list, package = "scRepertoire")
data <- scRepertoire::combineTCR(contig_list)
data <- scRepertoire::addVariable(data,
variable.name = "Type",
variables = sample(c("B", "L"), 8, replace = TRUE)
)
data <- scRepertoire::addVariable(data,
variable.name = "Sex",
variables = sample(c("M", "F"), 8, replace = TRUE)
)
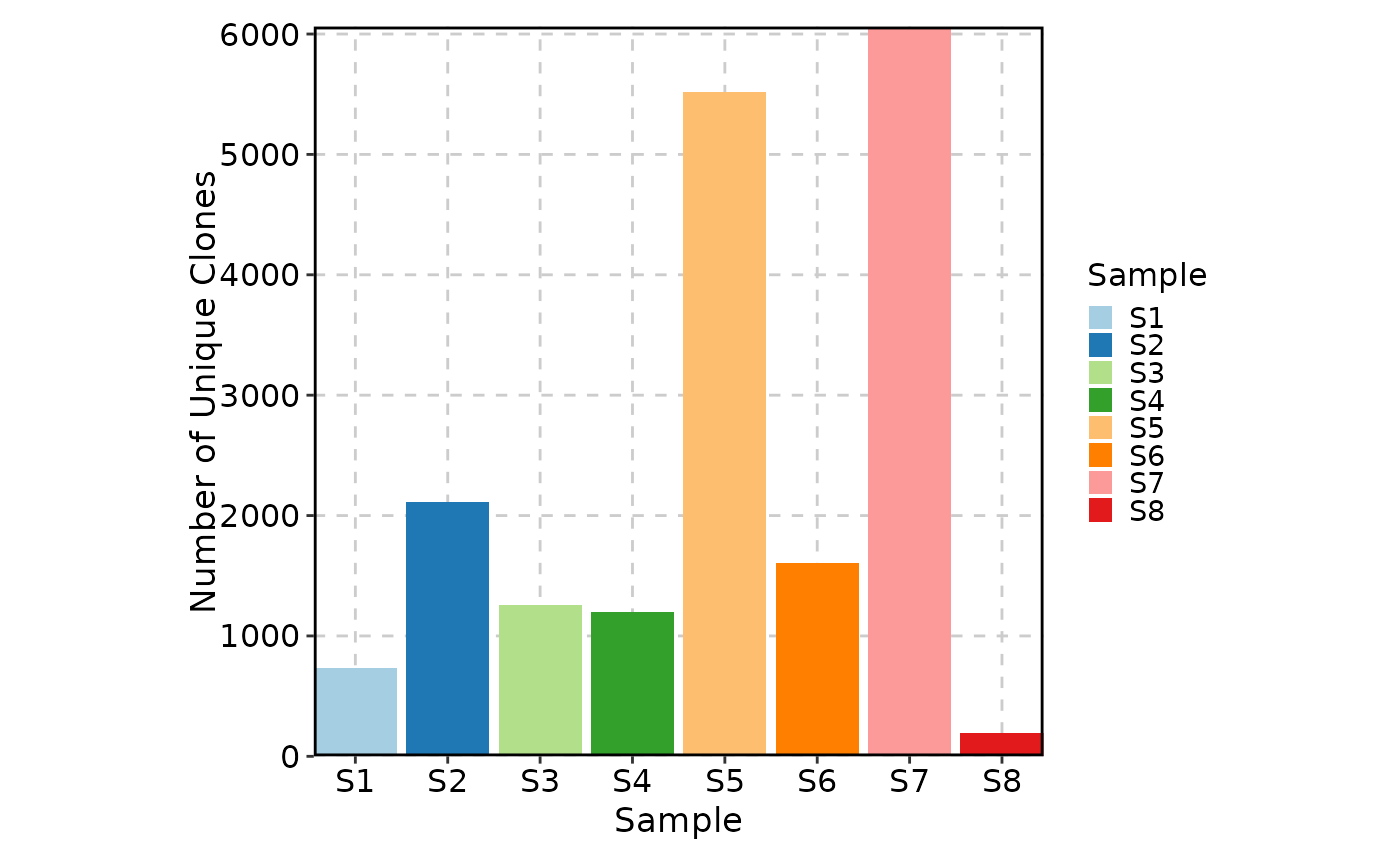
ClonalVolumePlot(data)
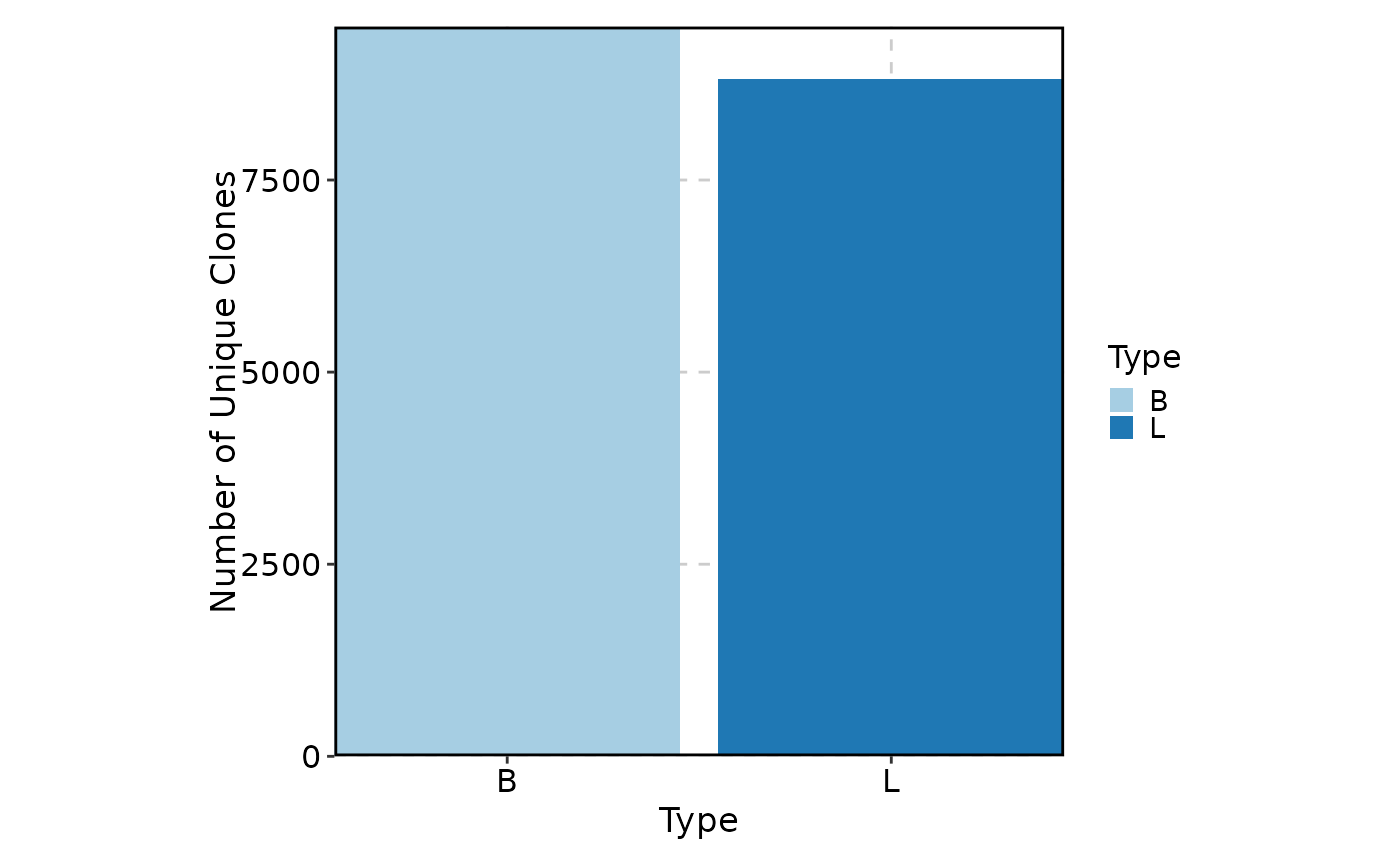
 ClonalVolumePlot(data, x = "Type")
ClonalVolumePlot(data, x = "Type")
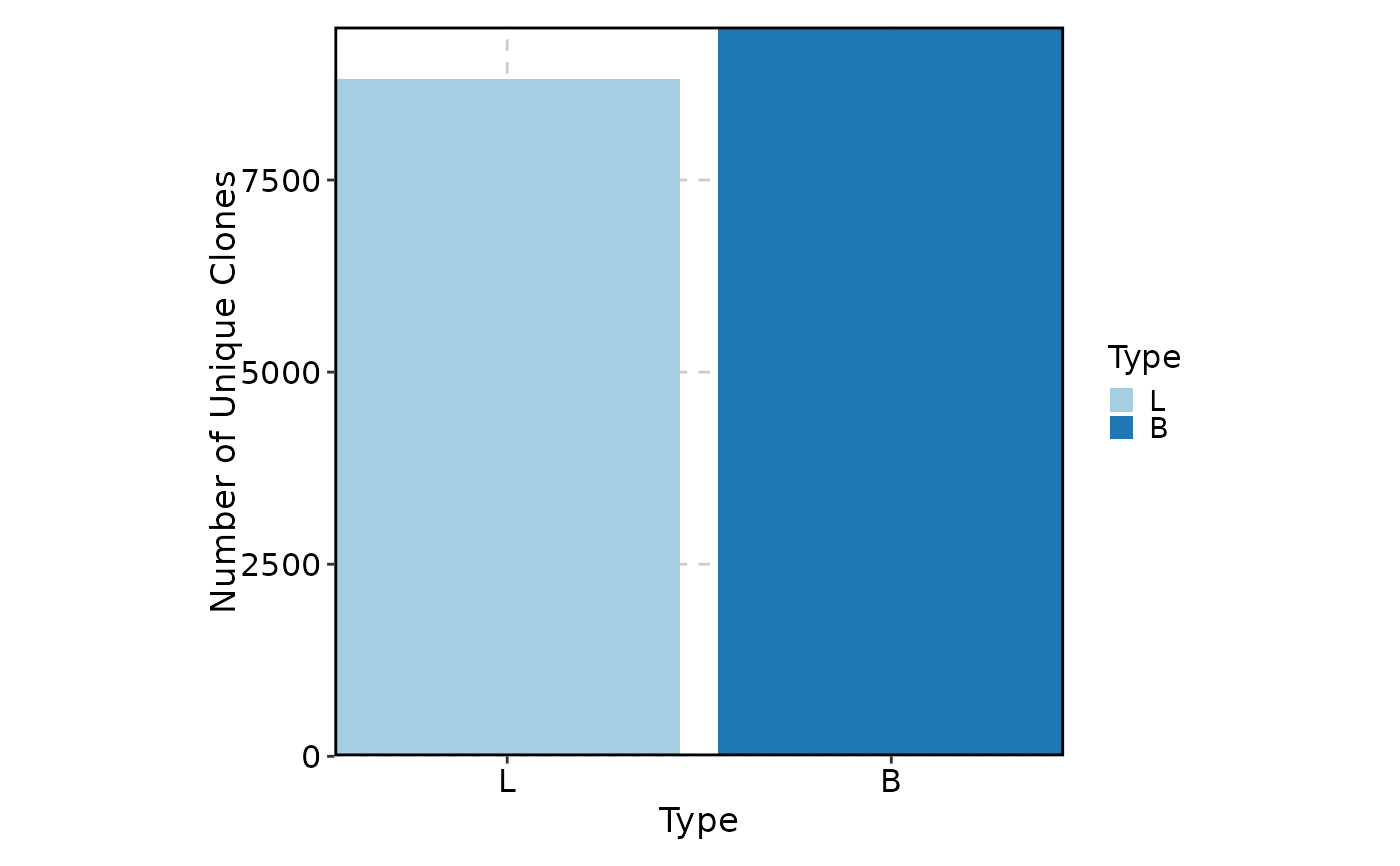
 ClonalVolumePlot(data, x = "Type", order = list(Type = c("L", "B")))
ClonalVolumePlot(data, x = "Type", order = list(Type = c("L", "B")))
 ClonalVolumePlot(data, x = c("Type", "Sex"), scale = TRUE)
#> Multiple columns are provided in 'x'. They will be concatenated into one column.
ClonalVolumePlot(data, x = c("Type", "Sex"), scale = TRUE)
#> Multiple columns are provided in 'x'. They will be concatenated into one column.
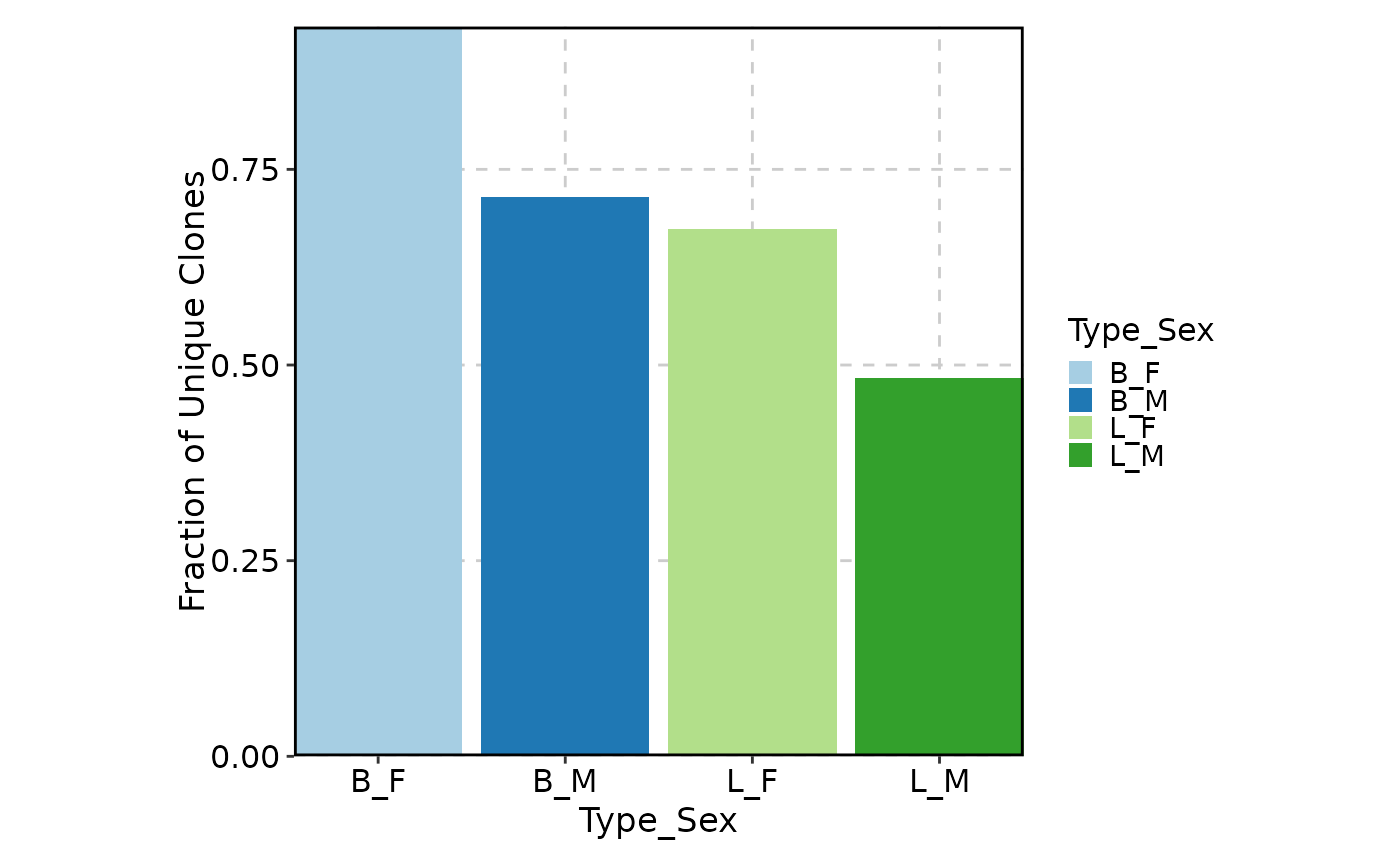 ClonalVolumePlot(data, x = "Type", group_by = "Sex", position = "stack")
ClonalVolumePlot(data, x = "Type", group_by = "Sex", position = "stack")
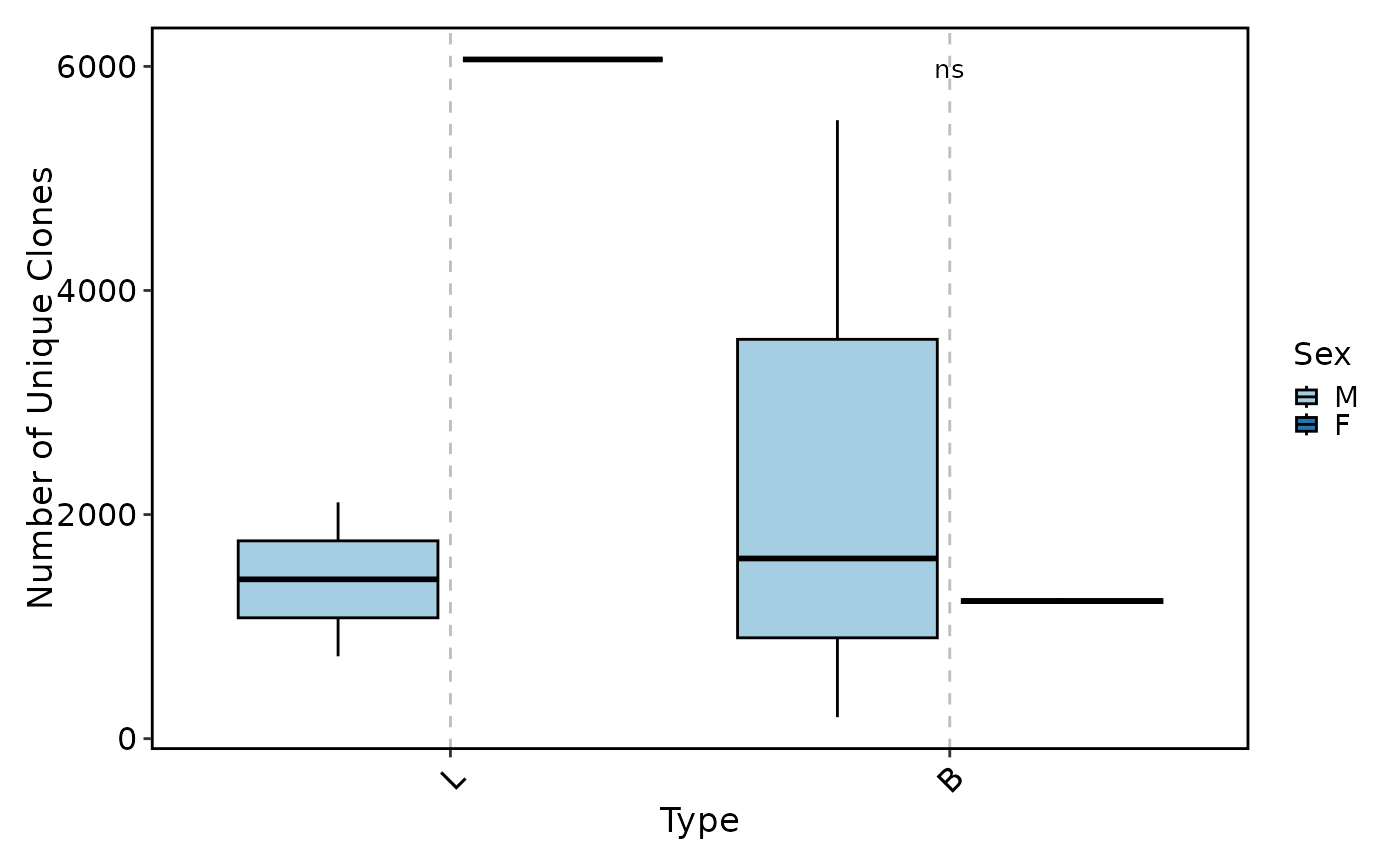
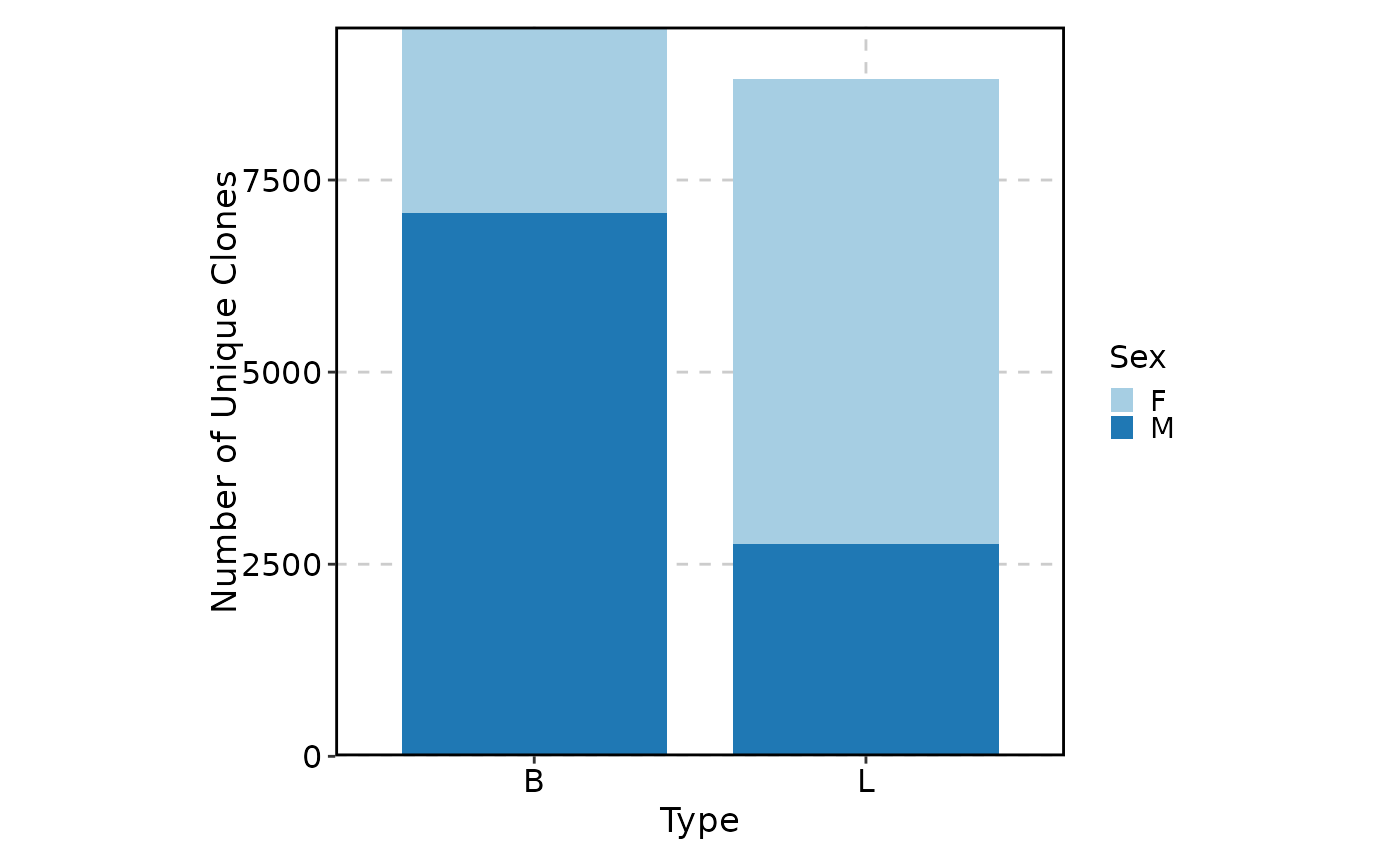 ClonalVolumePlot(data,
plot_type = "box", x = "Type", comparisons = TRUE,
group_by = "Sex"
)
ClonalVolumePlot(data,
plot_type = "box", x = "Type", comparisons = TRUE,
group_by = "Sex"
)
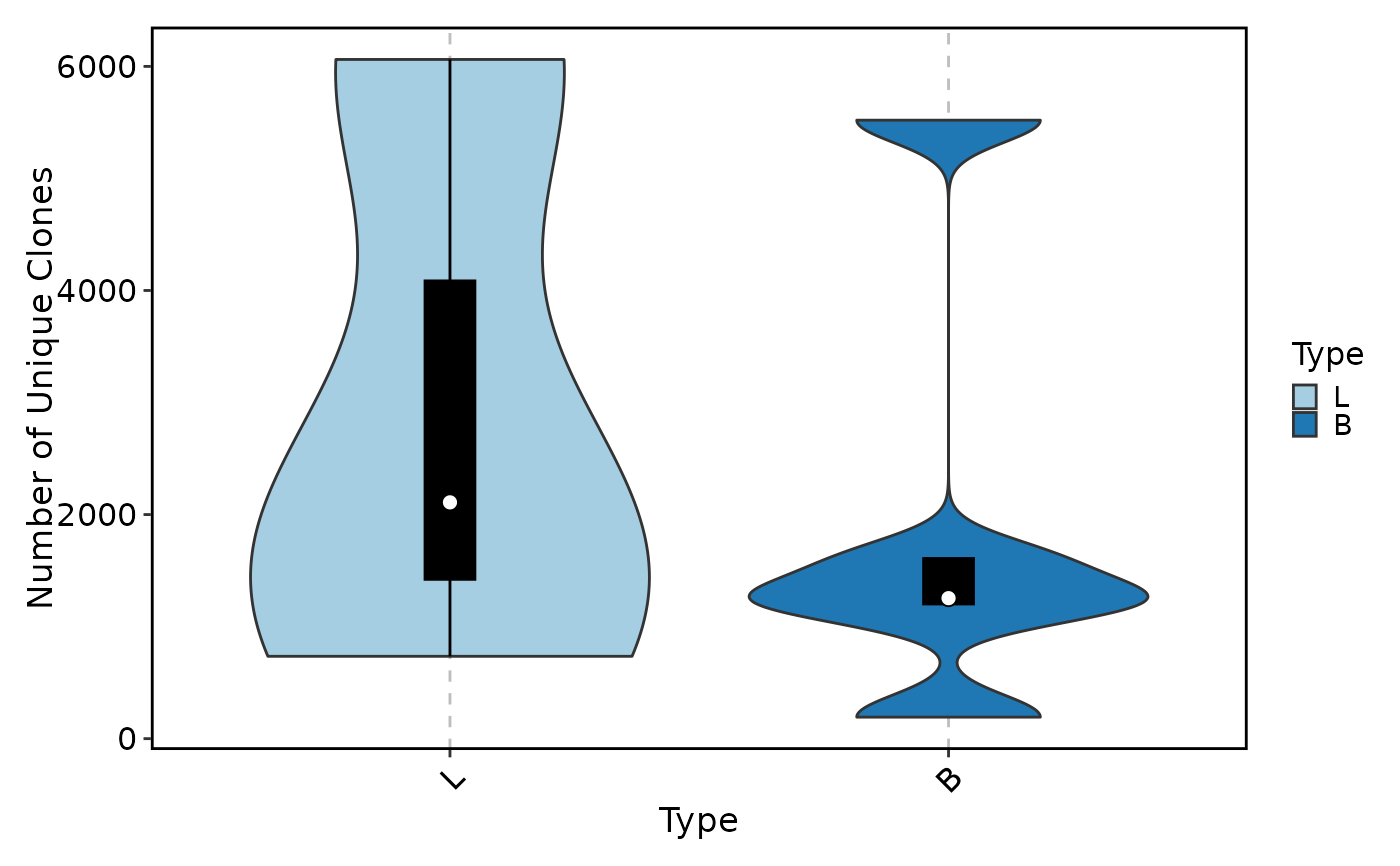
 ClonalVolumePlot(data, plot_type = "violin", x = "Type", add_box = TRUE)
ClonalVolumePlot(data, plot_type = "violin", x = "Type", add_box = TRUE)
 # on a Seurat object
data(scRep_example, package = "scRepertoire")
data(contig_list, package = "scRepertoire")
combined <- scRepertoire::combineTCR(contig_list,
samples = c("P17B", "P17L", "P18B", "P18L", "P19B", "P19L", "P20B", "P20L")
)
sobj <- scRepertoire::combineExpression(combined, scRep_example)
ClonalVolumePlot(sobj)
#> Warning: The 'Sample' column is not found in the meta data, 'orig.indent' will be used instead.
# on a Seurat object
data(scRep_example, package = "scRepertoire")
data(contig_list, package = "scRepertoire")
combined <- scRepertoire::combineTCR(contig_list,
samples = c("P17B", "P17L", "P18B", "P18L", "P19B", "P19L", "P20B", "P20L")
)
sobj <- scRepertoire::combineExpression(combined, scRep_example)
ClonalVolumePlot(sobj)
#> Warning: The 'Sample' column is not found in the meta data, 'orig.indent' will be used instead.
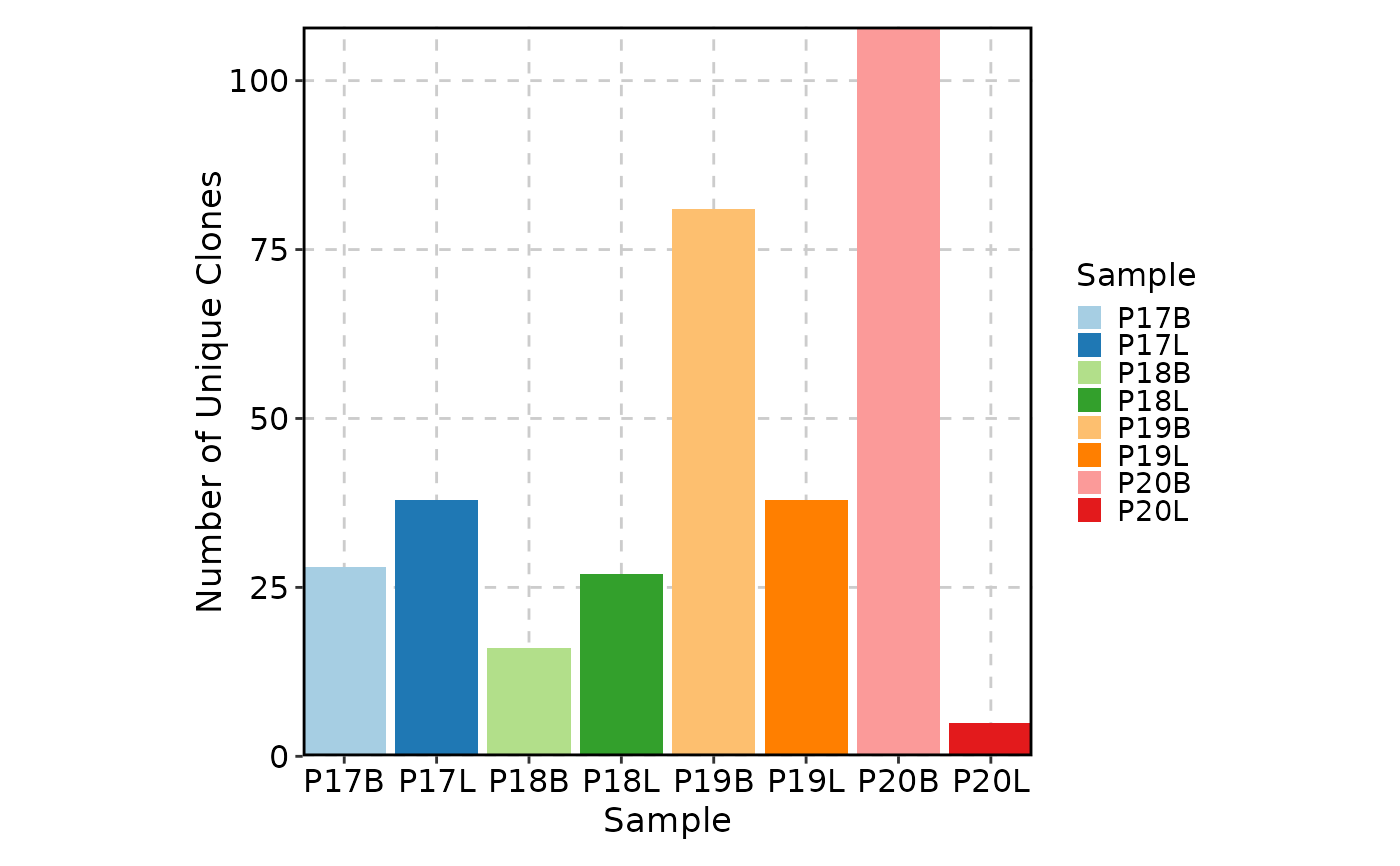 ClonalVolumePlot(sobj, x = "seurat_clusters")
#> Warning: The 'Sample' column is not found in the meta data, 'orig.indent' will be used instead.
ClonalVolumePlot(sobj, x = "seurat_clusters")
#> Warning: The 'Sample' column is not found in the meta data, 'orig.indent' will be used instead.
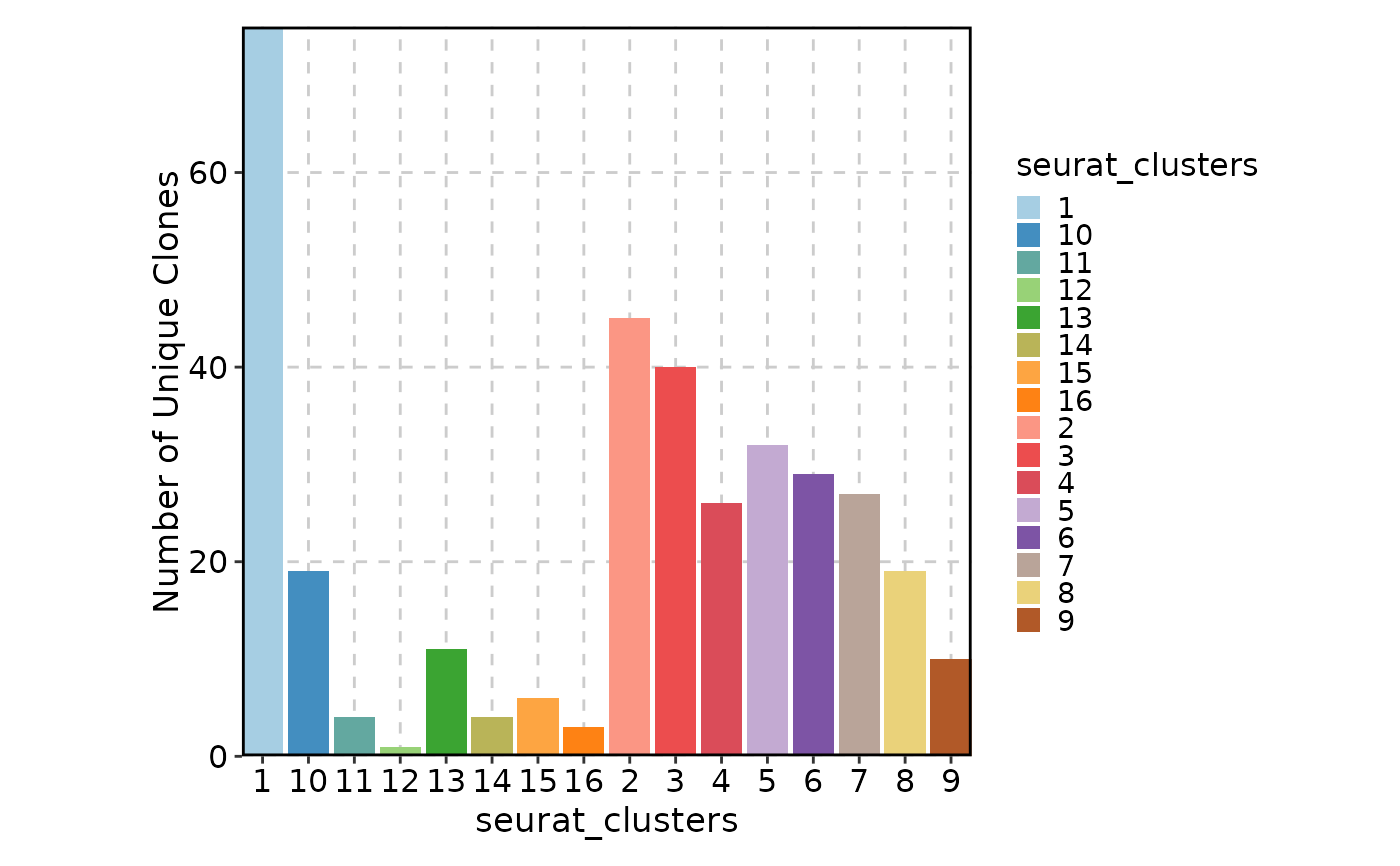 ClonalVolumePlot(sobj, group_by = "seurat_clusters")
#> Warning: The 'Sample' column is not found in the meta data, 'orig.indent' will be used instead.
ClonalVolumePlot(sobj, group_by = "seurat_clusters")
#> Warning: The 'Sample' column is not found in the meta data, 'orig.indent' will be used instead.
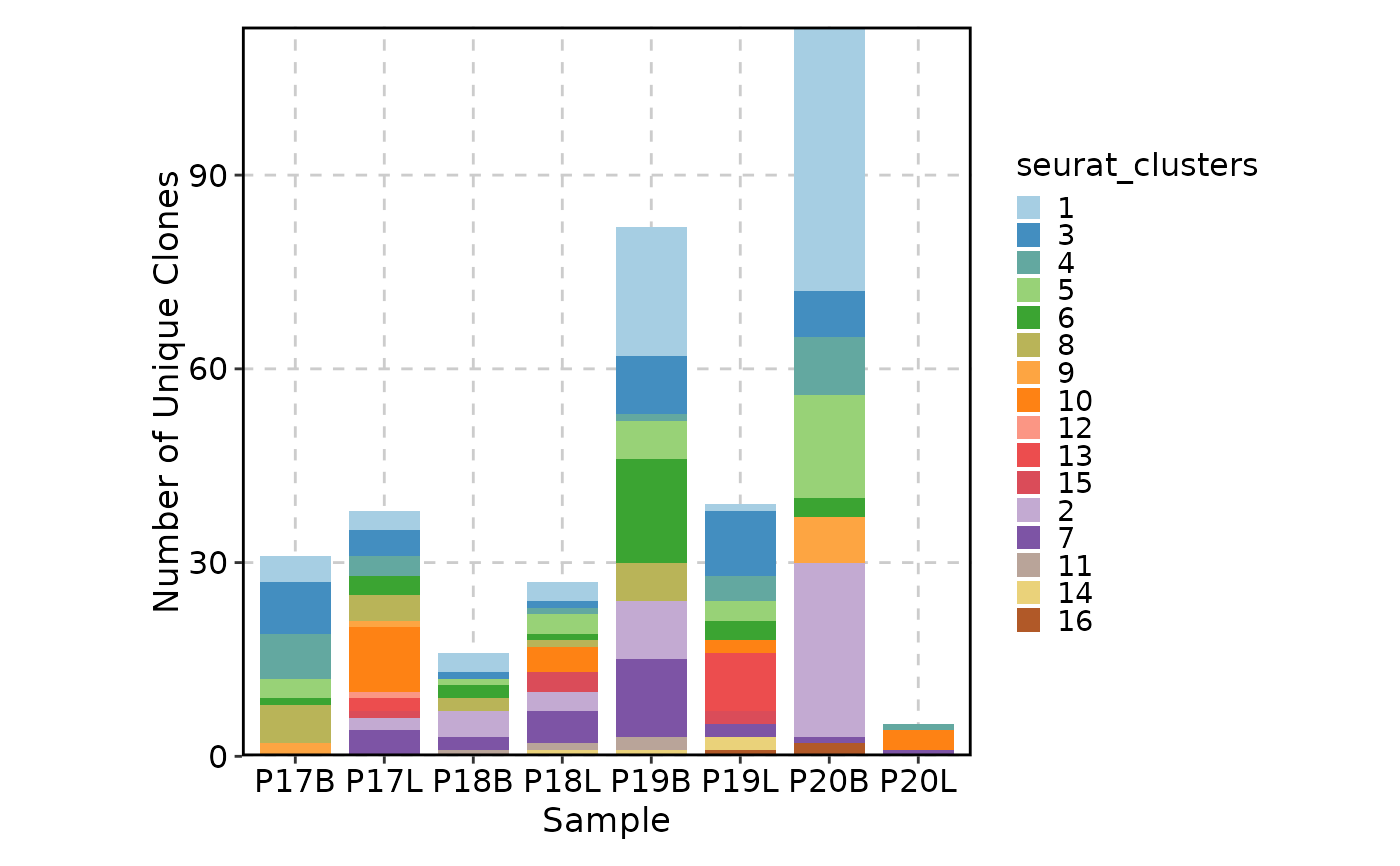 ClonalVolumePlot(sobj, x = "seurat_clusters", plot_type = "box")
#> Warning: The 'Sample' column is not found in the meta data, 'orig.indent' will be used instead.
ClonalVolumePlot(sobj, x = "seurat_clusters", plot_type = "box")
#> Warning: The 'Sample' column is not found in the meta data, 'orig.indent' will be used instead.
 # }
# }