Plot the rarefaction curves
Usage
ClonalRarefactionPlot(
data,
clone_call = "aa",
chain = "both",
group_by = "Sample",
group_by_sep = "_",
n_boots = 20,
q = 0,
facet_by = NULL,
split_by = NULL,
split_by_sep = "_",
palette = "Paired",
combine = TRUE,
nrow = NULL,
ncol = NULL,
byrow = TRUE,
...
)Arguments
- data
The product of scRepertoire::combineTCR, scRepertoire::combineTCR, or scRepertoire::combineExpression.
- clone_call
How to call the clone - VDJC gene (gene), CDR3 nucleotide (nt), CDR3 amino acid (aa), VDJC gene + CDR3 nucleotide (strict) or a custom variable
- chain
indicate if both or a specific chain should be used - e.g. "both", "TRA", "TRG", "IGH", "IGL"
- group_by
A character vector of column names to group the samples. Default is "Sample".
- group_by_sep
The separator for the group_by column. Default is "_".
- n_boots
The number of bootstrap samples. Default is 20.
- q
The hill number. Default is 0.
0 - Species richness
1 - Shannon entropy
2 - Simpson index#'
- facet_by
A character vector of column names to facet the plots. Default is NULL.
- split_by
A character vector of column names to split the plots. Default is NULL.
- split_by_sep
The separator for the split_by column. Default is "_".
- palette
The color palette to use. Default is "Paired".
- combine
Whether to combine the plots into a single plot. Default is TRUE.
- nrow
The number of rows in the combined plot. Default is NULL.
- ncol
The number of columns in the combined plot. Default is NULL.
- byrow
Whether to fill the combined plot by row. Default is TRUE.
- ...
Other arguments passed to
plotthis::RarefactionPlot().
Examples
# \donttest{
set.seed(8525)
data(contig_list, package = "scRepertoire")
data <- scRepertoire::combineTCR(contig_list,
samples = c("P17B", "P17L", "P18B", "P18L", "P19B","P19L", "P20B", "P20L"))
data <- scRepertoire::addVariable(data,
variable.name = "Type",
variables = rep(c("B", "L"), 4)
)
data <- scRepertoire::addVariable(data,
variable.name = "Subject",
variables = rep(c("P17", "P18", "P19", "P20"), each = 2)
)
ClonalRarefactionPlot(data, type = 1, q = 0, n_boots = 2)
#> Warning: The shape palette can deal with a maximum of 6 discrete values because more
#> than 6 becomes difficult to discriminate
#> ℹ you have requested 8 values. Consider specifying shapes manually if you need
#> that many of them.
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_point()`).
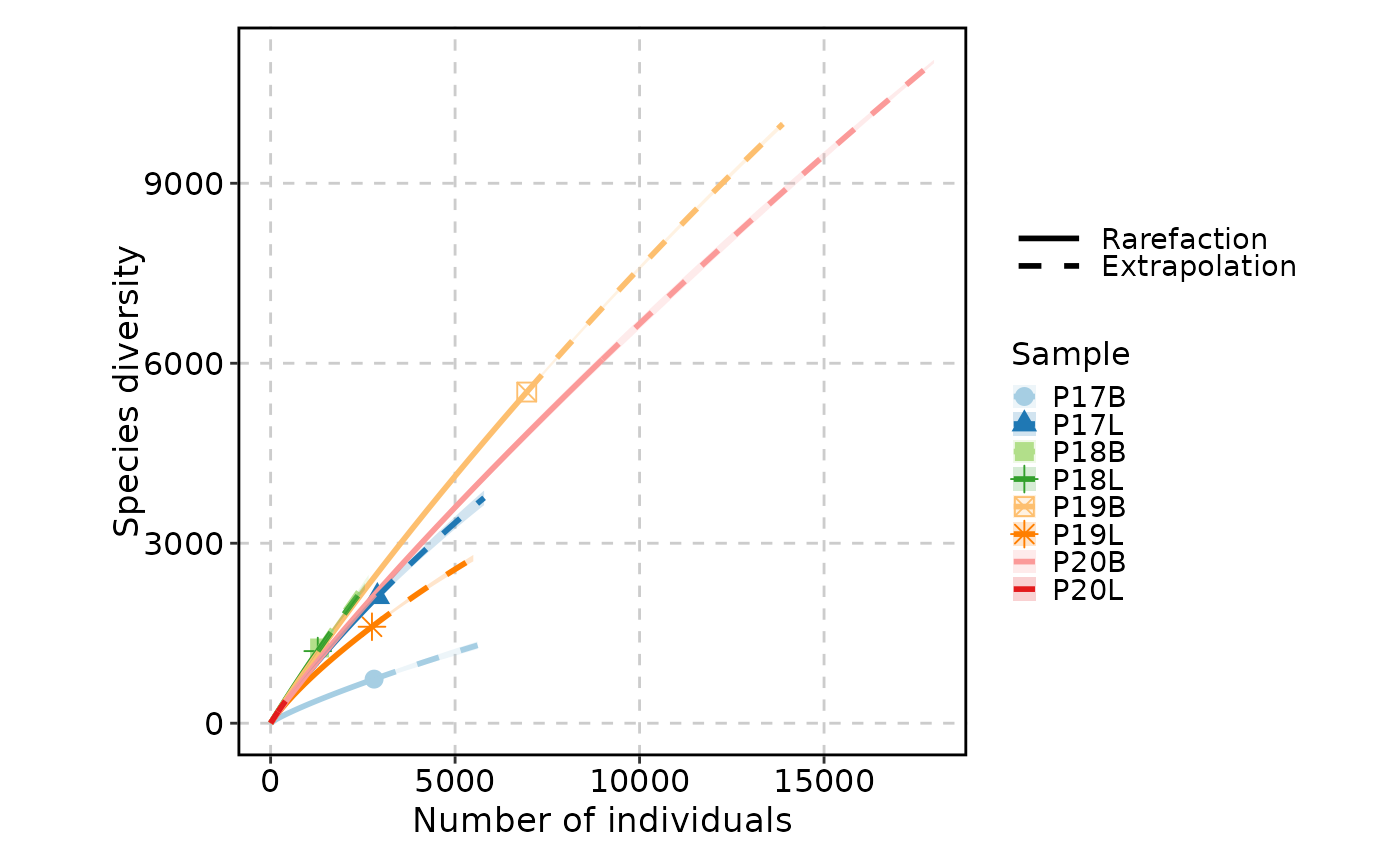 ClonalRarefactionPlot(data, type = 2, q = 0, n_boots = 2)
#> Warning: The shape palette can deal with a maximum of 6 discrete values because more
#> than 6 becomes difficult to discriminate
#> ℹ you have requested 8 values. Consider specifying shapes manually if you need
#> that many of them.
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_point()`).
ClonalRarefactionPlot(data, type = 2, q = 0, n_boots = 2)
#> Warning: The shape palette can deal with a maximum of 6 discrete values because more
#> than 6 becomes difficult to discriminate
#> ℹ you have requested 8 values. Consider specifying shapes manually if you need
#> that many of them.
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_point()`).
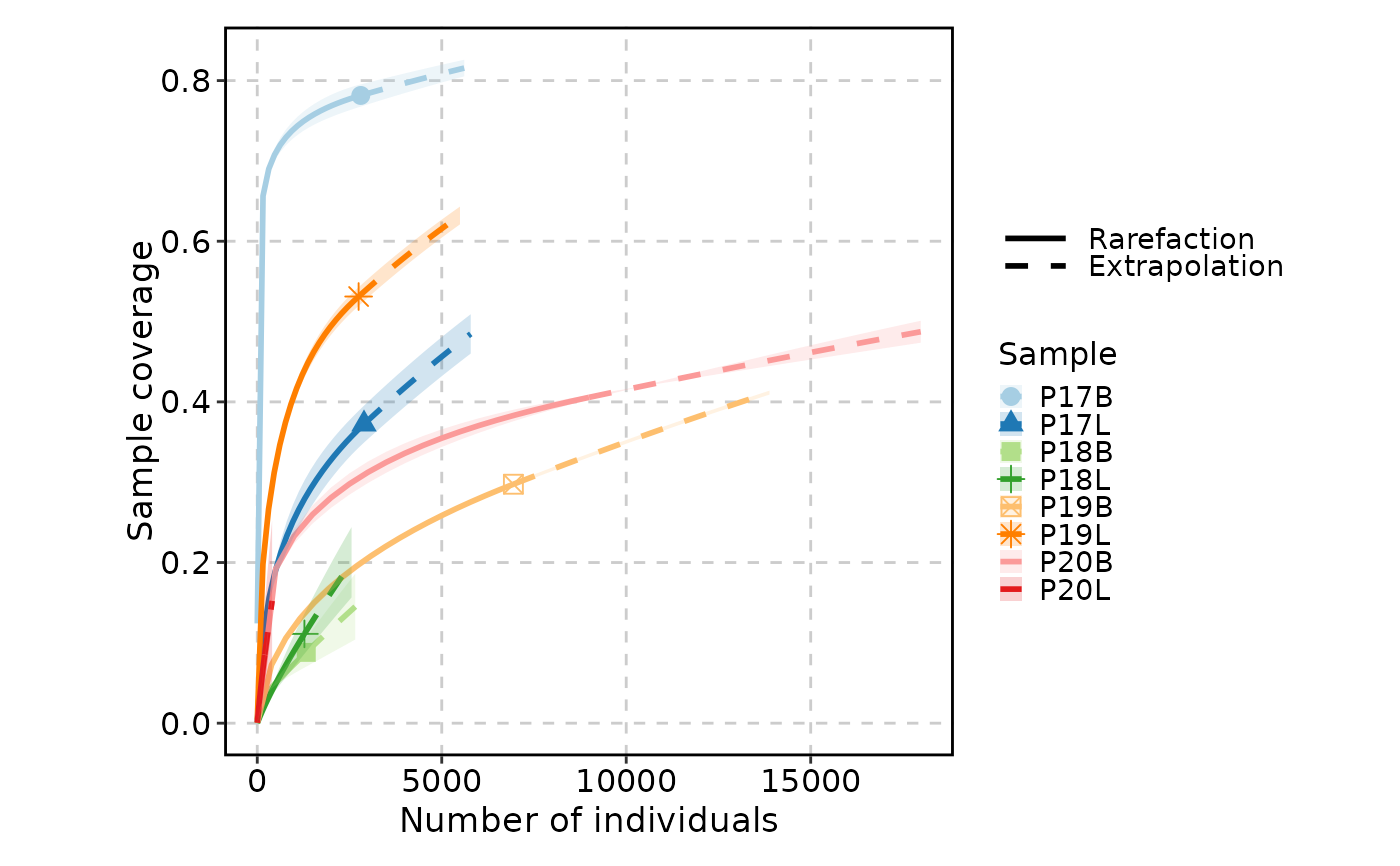 ClonalRarefactionPlot(data, type = 3, q = 0, n_boots = 2)
#> Warning: The shape palette can deal with a maximum of 6 discrete values because more
#> than 6 becomes difficult to discriminate
#> ℹ you have requested 8 values. Consider specifying shapes manually if you need
#> that many of them.
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_point()`).
ClonalRarefactionPlot(data, type = 3, q = 0, n_boots = 2)
#> Warning: The shape palette can deal with a maximum of 6 discrete values because more
#> than 6 becomes difficult to discriminate
#> ℹ you have requested 8 values. Consider specifying shapes manually if you need
#> that many of them.
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_point()`).
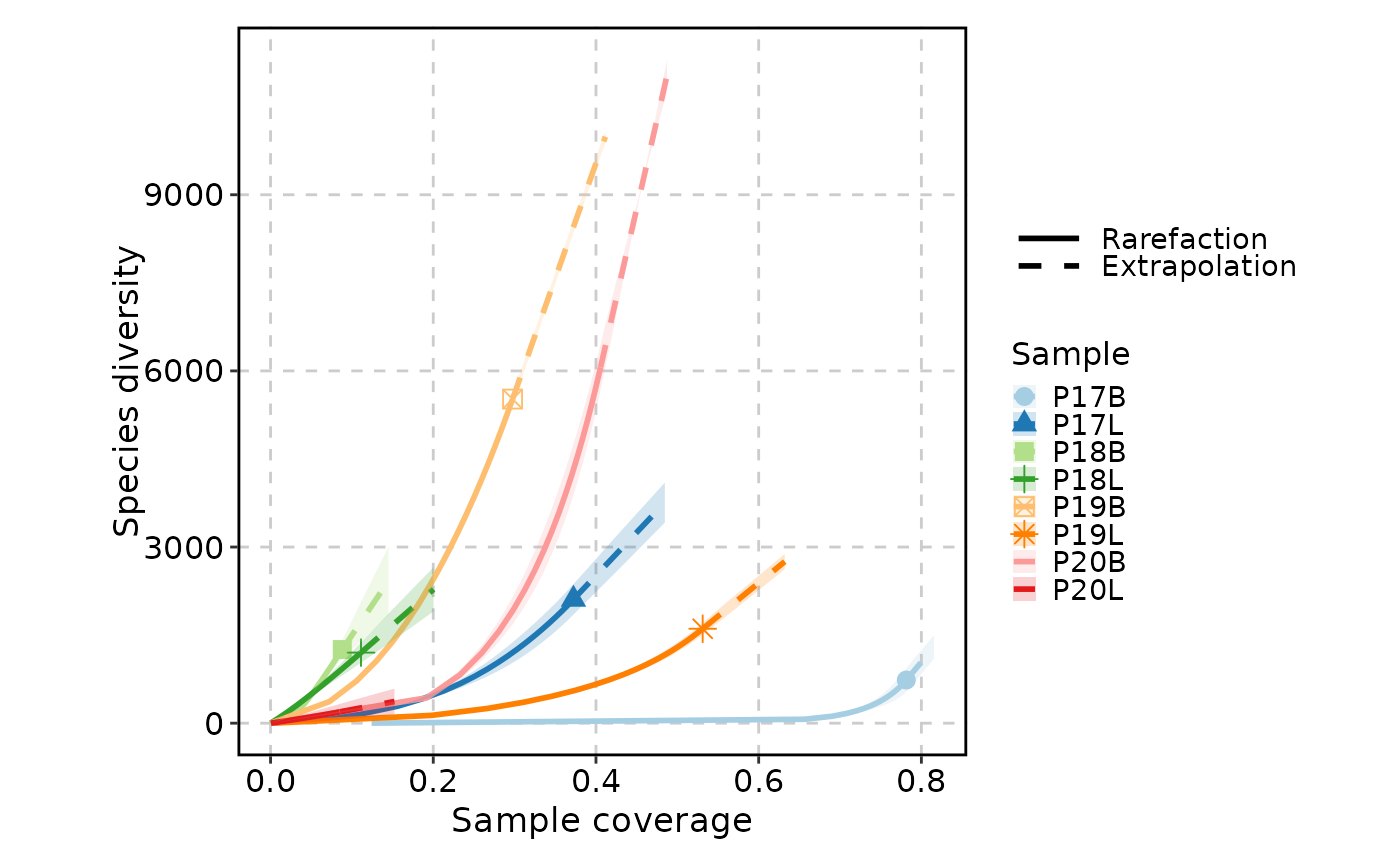 ClonalRarefactionPlot(data, q = 1, n_boots = 2)
#> Warning: The shape palette can deal with a maximum of 6 discrete values because more
#> than 6 becomes difficult to discriminate
#> ℹ you have requested 8 values. Consider specifying shapes manually if you need
#> that many of them.
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_point()`).
ClonalRarefactionPlot(data, q = 1, n_boots = 2)
#> Warning: The shape palette can deal with a maximum of 6 discrete values because more
#> than 6 becomes difficult to discriminate
#> ℹ you have requested 8 values. Consider specifying shapes manually if you need
#> that many of them.
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_point()`).
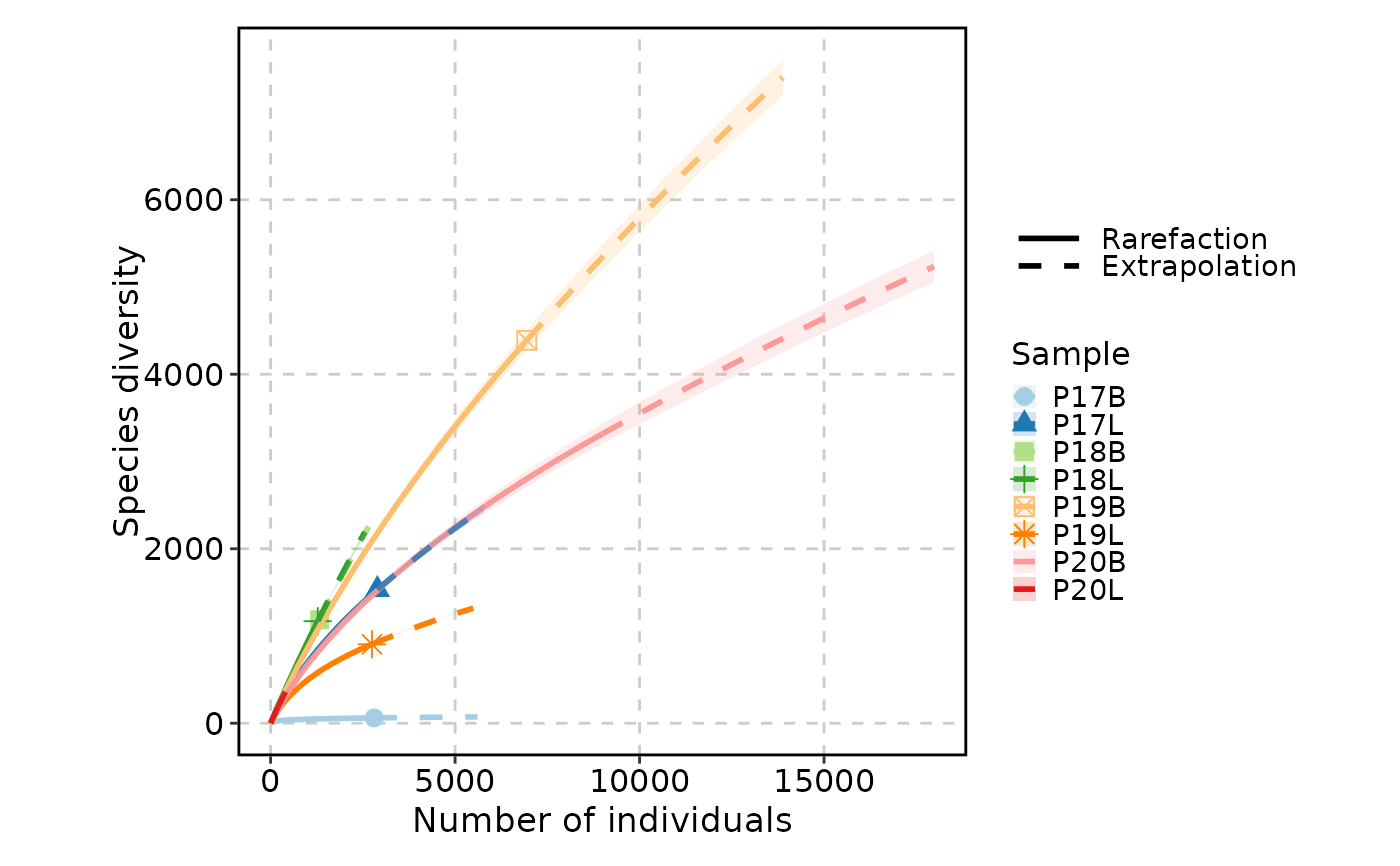 ClonalRarefactionPlot(data, q = 1, n_boots = 2, group_by = "Type")
ClonalRarefactionPlot(data, q = 1, n_boots = 2, group_by = "Type")
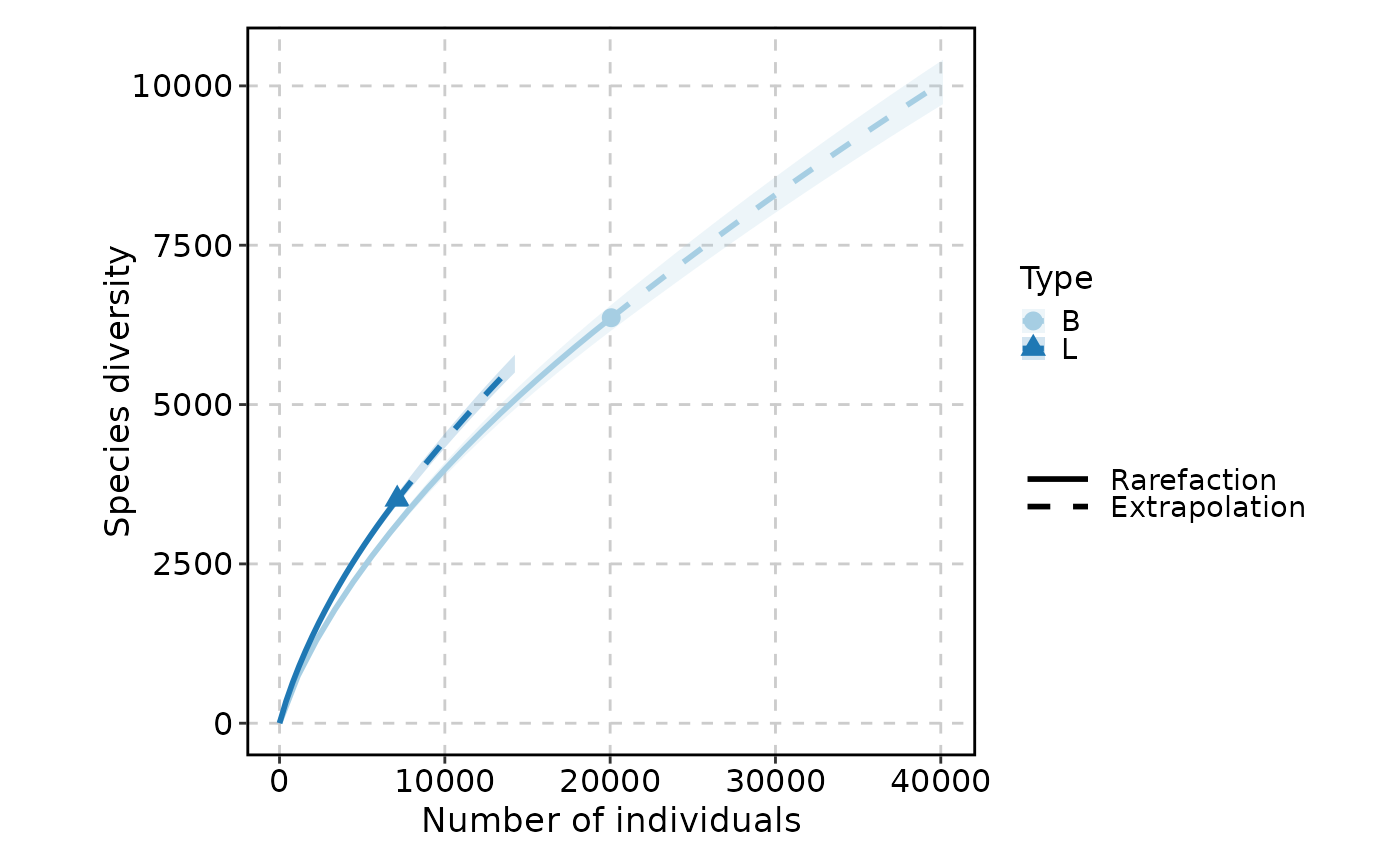 ClonalRarefactionPlot(data, n_boots = 2, split_by = "Type")
ClonalRarefactionPlot(data, n_boots = 2, split_by = "Type")
 # }
# }