This function creates various types of feature statistic plots for a Seurat object, a Giotto object,
a path to an .h5ad file or an opened H5File by hdf5r package.
It allows for plotting features such as gene expression, scores, or other metadata across different groups or conditions.
The function supports multiple plot types including violin, box, bar, ridge, dimension reduction, correlation, heatmap, and dot plots.
It can also handle multiple features and supports faceting, splitting, and grouping by metadata columns.
Usage
FeatureStatPlot(
object,
features,
plot_type = c("violin", "box", "bar", "ridge", "dim", "cor", "heatmap", "dot"),
spat_unit = NULL,
feat_type = NULL,
downsample = NULL,
pos_only = c("no", "any", "all"),
reduction = NULL,
graph = NULL,
bg_cutoff = 0,
dims = 1:2,
rows_name = "Features",
ident = NULL,
assay = NULL,
layer = NULL,
agg = mean,
group_by = NULL,
split_by = NULL,
facet_by = NULL,
xlab = NULL,
ylab = NULL,
x_text_angle = NULL,
...
)Arguments
- object
A seurat object, a giotto object, a path to an .h5ad file or an opened
H5Filebyhdf5rpackage.- features
A character vector of feature names
- plot_type
Type of the plot. It can be "violin", "box", "bar", "ridge", "dim", "cor", "heatmap" or "dot"
- spat_unit
The spatial unit to use for the plot. Only applied to Giotto objects.
- feat_type
feature type of the features (e.g. "rna", "dna", "protein"), only applied to Giotto objects.
- downsample
A numeric the number of cells in each identity group to downsample to for violin, box, or ridge plots. If n > 1, it is treated as the number of cells to downsample to. If 0 < n <= 1, it is treated as the fraction of cells to downsample to.
- pos_only
Whether to only include cells with positive feature values.
"no": Do not filter cells based on feature values. (default)
"any": Include cells with positive values for any of the features.
"all": Include cells with positive values for all of the features. If you have named features (i.e. a named list),
pos_onlywill be applied to all flattened features.
- reduction
Name of the reduction to plot (for example, "umap"), only used when
plot_typeis "dim" or you can to use the reduction as feature.- graph
Specify the graph name to add edges between cell neighbors to the plot, only used when
plot_typeis "dim".- bg_cutoff
Background cutoff for the dim plot, only used when
plot_typeis "dim".- dims
Dimensions to plot, only used when
plot_typeis "dim".- rows_name
The name of the rows in the heatmap, only used when
plot_typeis "heatmap".- ident
The column name in the meta data to identify the cells.
- assay
The assay to use for the feature data.
- layer
The layer to use for the feature data.
- agg
The aggregation function to use for the bar plot.
- group_by
The column name in the meta data to group the cells.
- split_by
Column name in the meta data to split the cells to different plots. If TRUE, the cells are split by the features.
- facet_by
Column name in the meta data to facet the plots. Should be always NULL.
- xlab
The x-axis label.
- ylab
The y-axis label.
- x_text_angle
The angle of the x-axis text. Only used when
plot_typeis "violin", "bar", or "box".- ...
Other arguments passed to the plot functions.
For
plot_type"violin", the arguments are passed toplotthis::ViolinPlot().For
plot_type"box", the arguments are passed toplotthis::BoxPlot().For
plot_type"bar", the arguments are passed toplotthis::BarPlot().For
plot_type"ridge", the arguments are passed toplotthis::RidgePlot().For
plot_type"dim", the arguments are passed toplotthis::FeatureDimPlot().For
plot_type"heatmap", the arguments are passed toplotthis::Heatmap().For
plot_type"cor" with 2 features, the arguments are passed toplotthis::CorPlot().For
plot_type"cor" with more than 2 features, the arguments are passed toplotthis::CorPairsPlot().For
plot_type"dot", the arguments are passed toplotthis::Heatmap()withcell_typeset to "dot".
Details
See:
https://pwwang.github.io/scplotter/articles/Giotto_Visium.html
https://pwwang.github.io/scplotter/articles/Giotto_Xenium.html
for examples of using this function with Giotto objects.
And see:
for examples of using this function with .h5ad files.
Examples
# \donttest{
data(pancreas_sub)
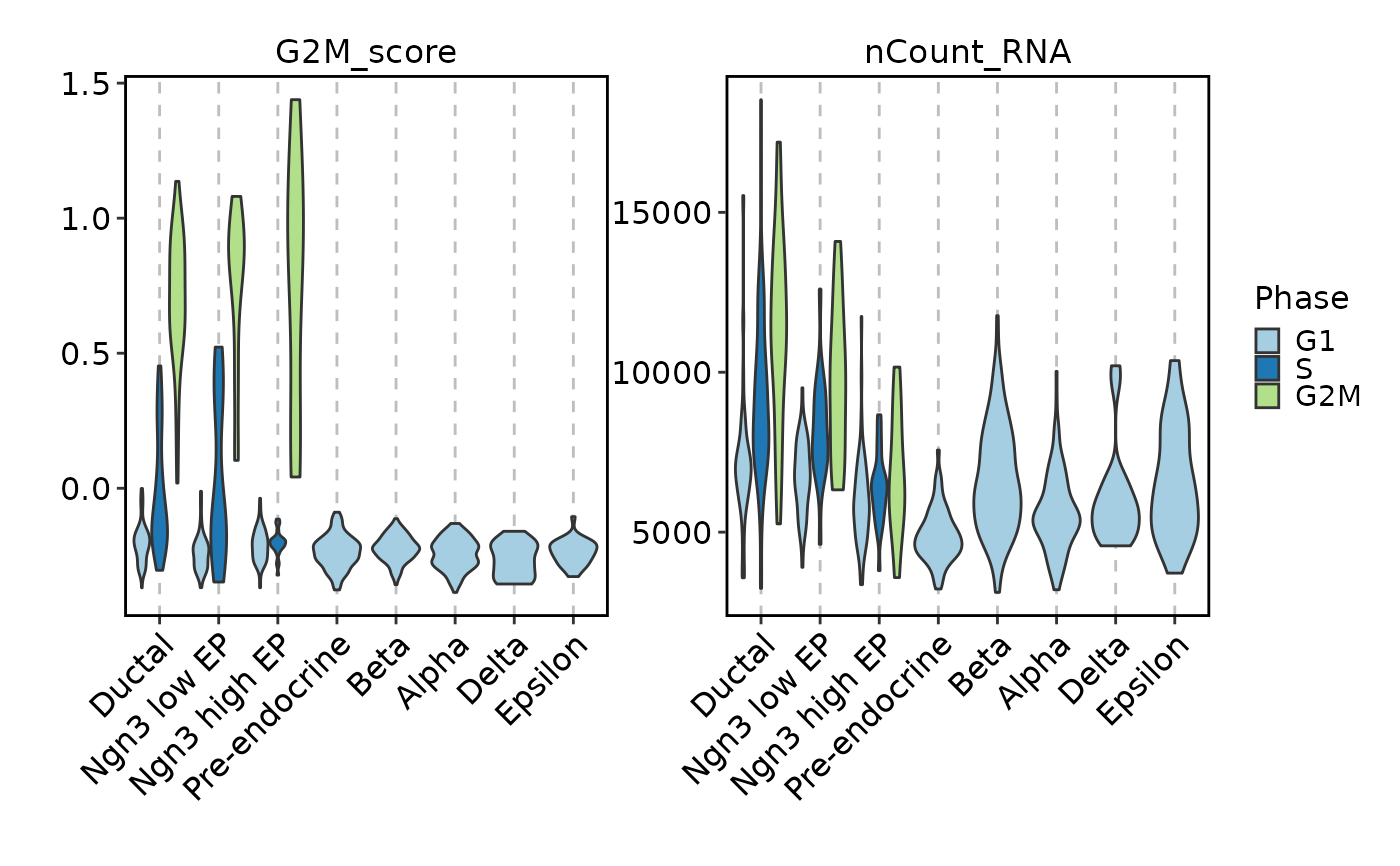
FeatureStatPlot(pancreas_sub, features = c("G2M_score", "nCount_RNA"),
ident = "SubCellType", facet_scales = "free_y")
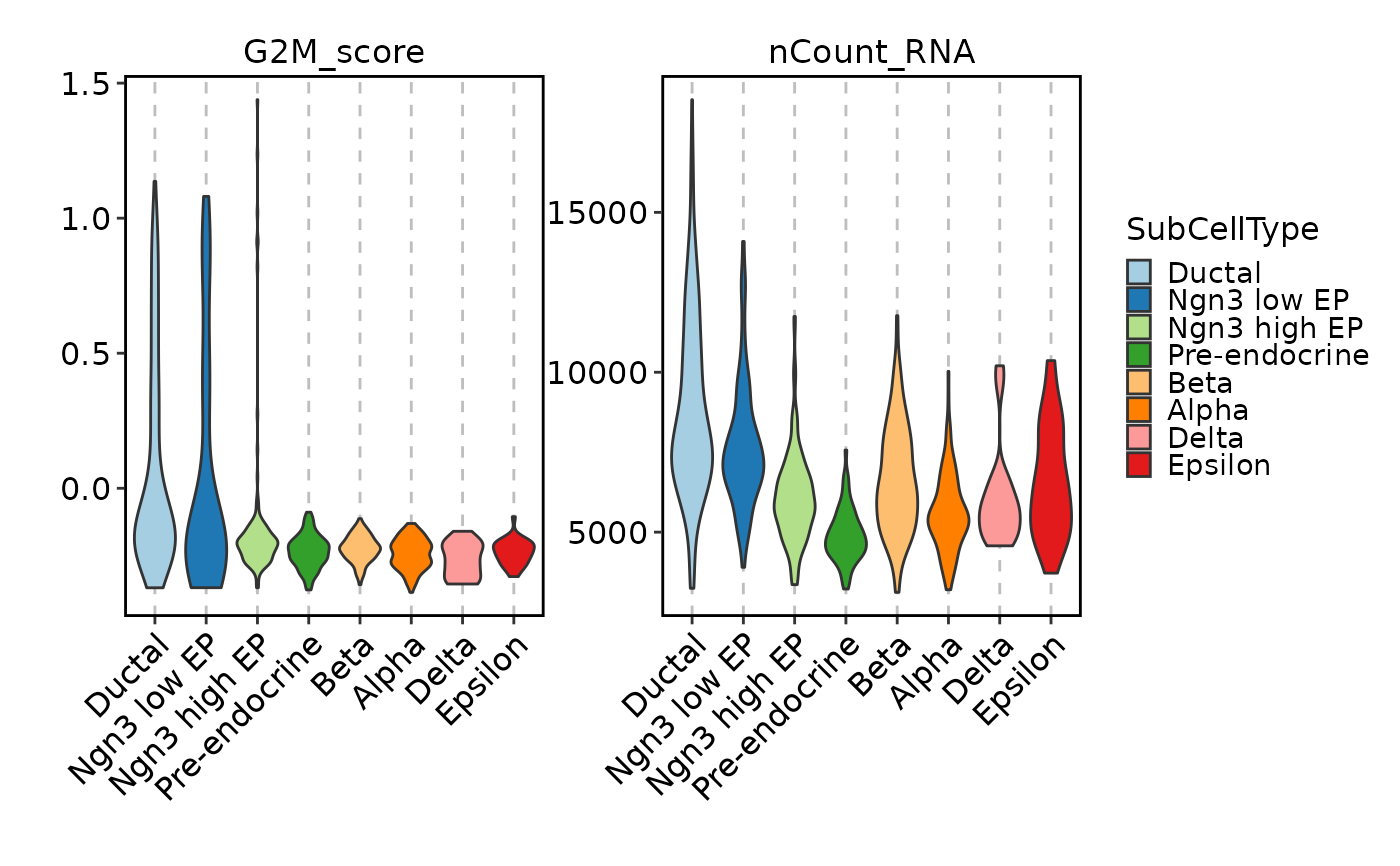 FeatureStatPlot(pancreas_sub, features = c("G2M_score", "nCount_RNA"),
ident = "SubCellType", plot_type = "box", facet_scales = "free_y")
FeatureStatPlot(pancreas_sub, features = c("G2M_score", "nCount_RNA"),
ident = "SubCellType", plot_type = "box", facet_scales = "free_y")
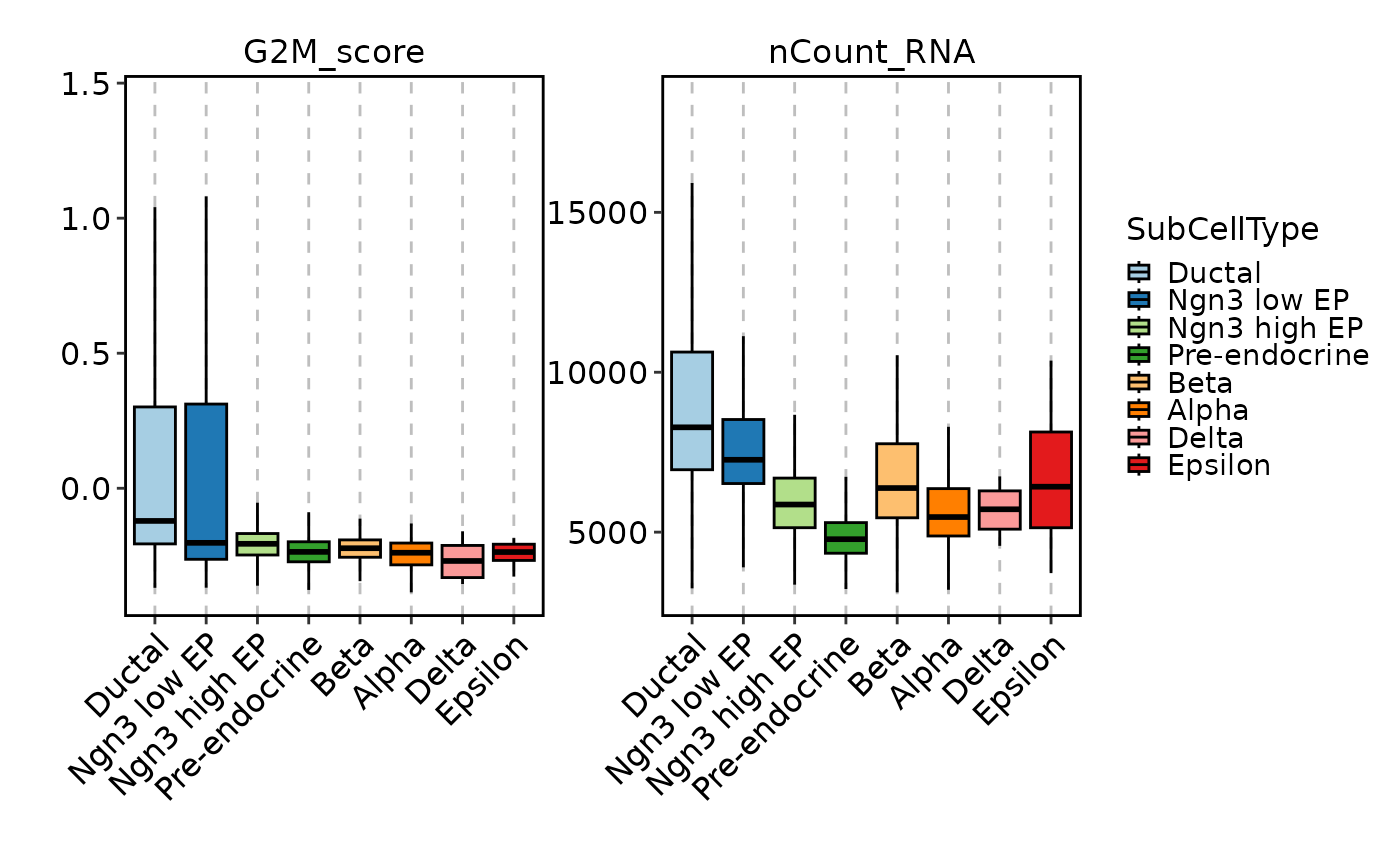 FeatureStatPlot(pancreas_sub, features = c("G2M_score", "nCount_RNA"),
ident = "SubCellType", plot_type = "bar", facet_scales = "free_y")
FeatureStatPlot(pancreas_sub, features = c("G2M_score", "nCount_RNA"),
ident = "SubCellType", plot_type = "bar", facet_scales = "free_y")
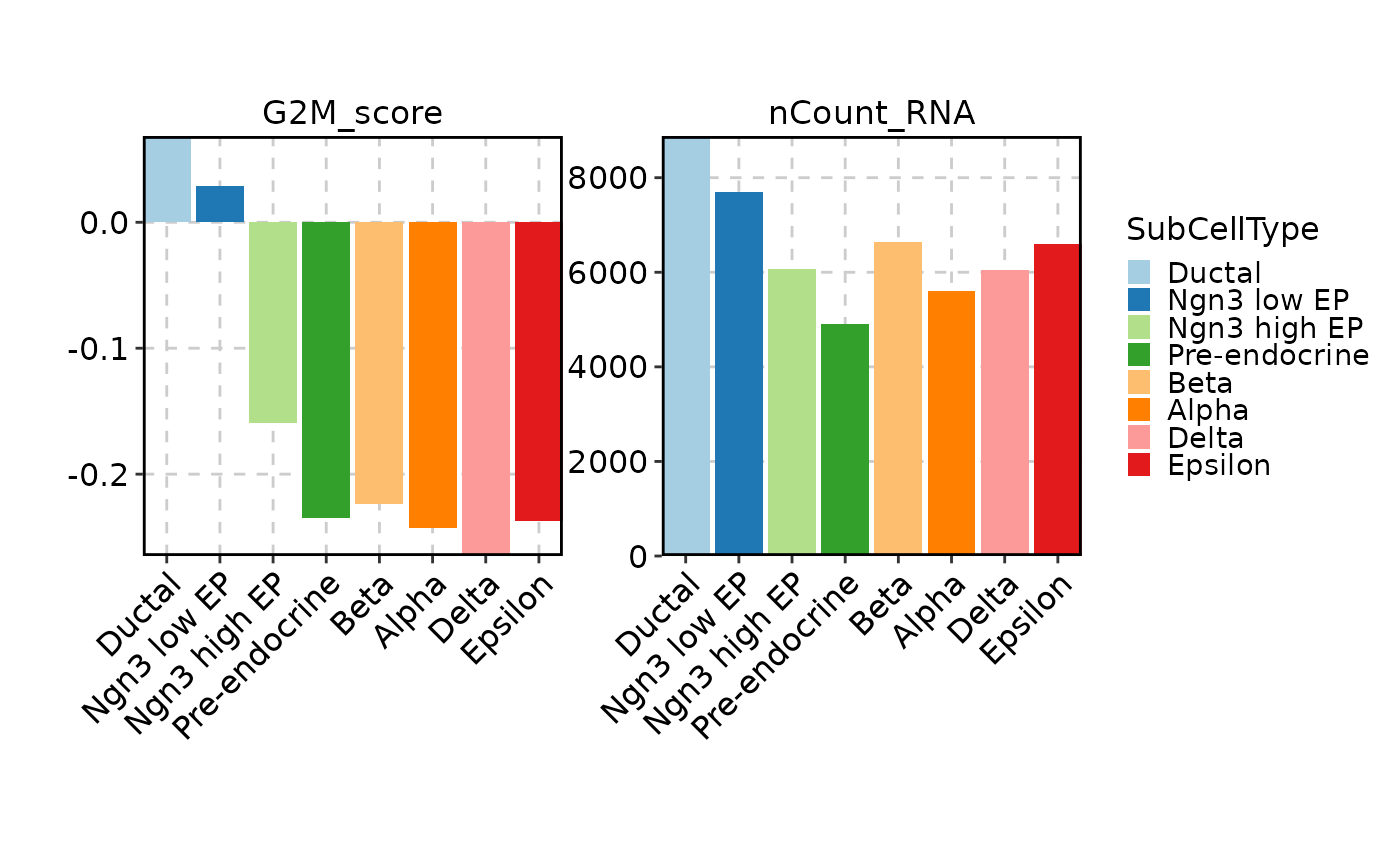 FeatureStatPlot(pancreas_sub, features = c("G2M_score", "nCount_RNA"),
ident = "SubCellType", plot_type = "ridge", flip = TRUE, facet_scales = "free_y")
#> Picking joint bandwidth of 0.0498
#> Picking joint bandwidth of 516
#> Picking joint bandwidth of 0.0498
#> Picking joint bandwidth of 516
FeatureStatPlot(pancreas_sub, features = c("G2M_score", "nCount_RNA"),
ident = "SubCellType", plot_type = "ridge", flip = TRUE, facet_scales = "free_y")
#> Picking joint bandwidth of 0.0498
#> Picking joint bandwidth of 516
#> Picking joint bandwidth of 0.0498
#> Picking joint bandwidth of 516
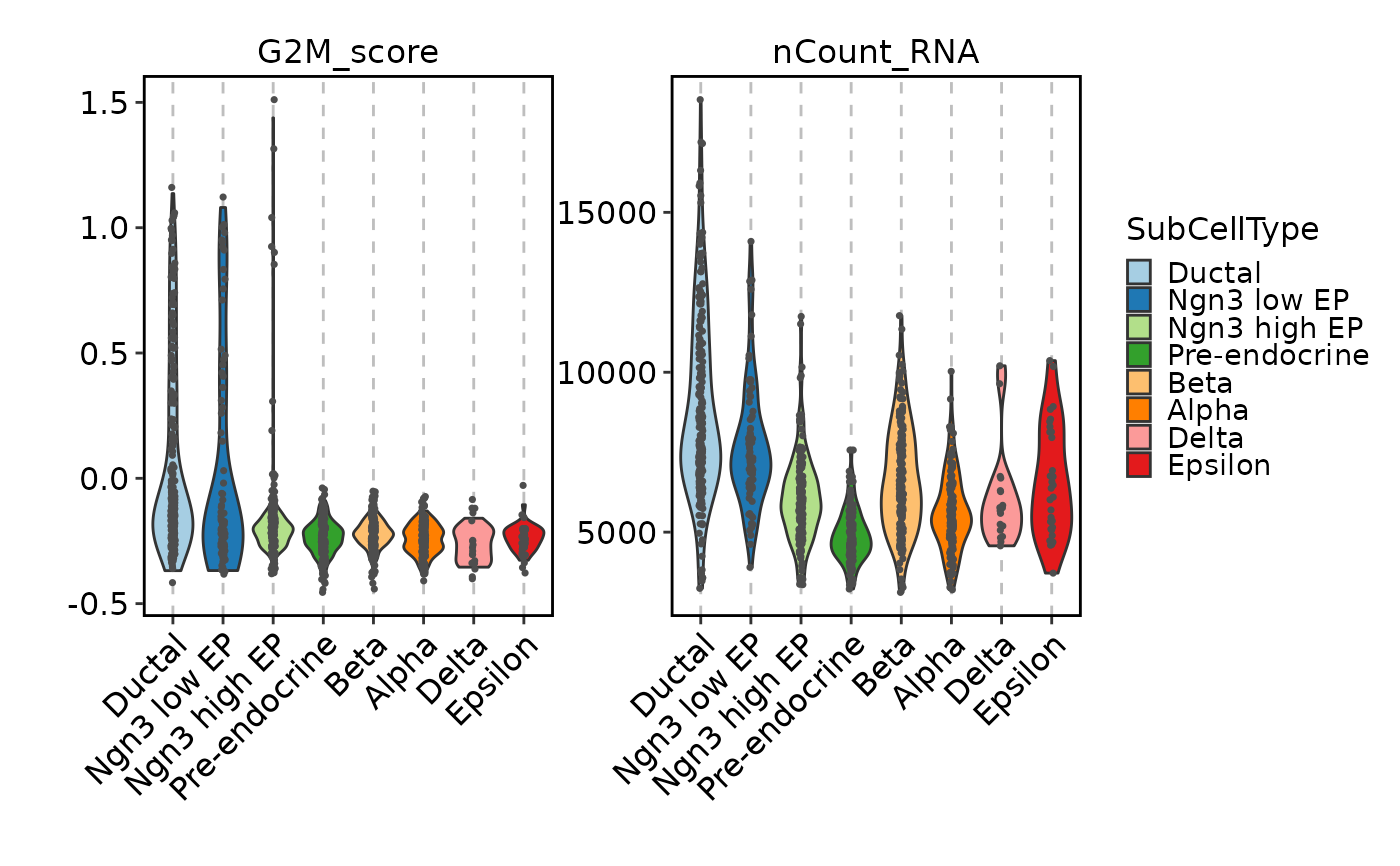
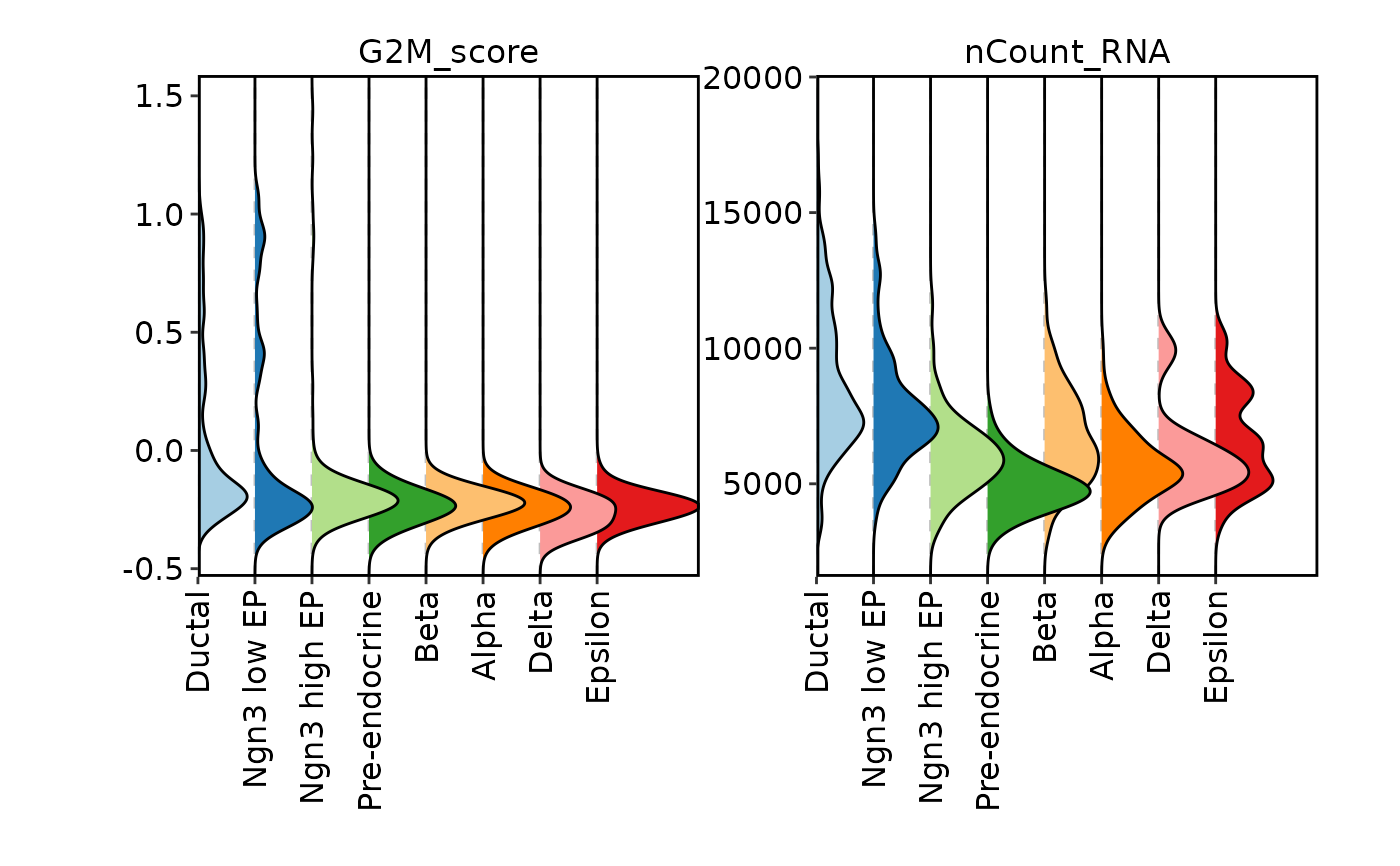 FeatureStatPlot(pancreas_sub, features = c("G2M_score", "nCount_RNA"),
ident = "SubCellType", facet_scales = "free_y", add_point = TRUE)
FeatureStatPlot(pancreas_sub, features = c("G2M_score", "nCount_RNA"),
ident = "SubCellType", facet_scales = "free_y", add_point = TRUE)
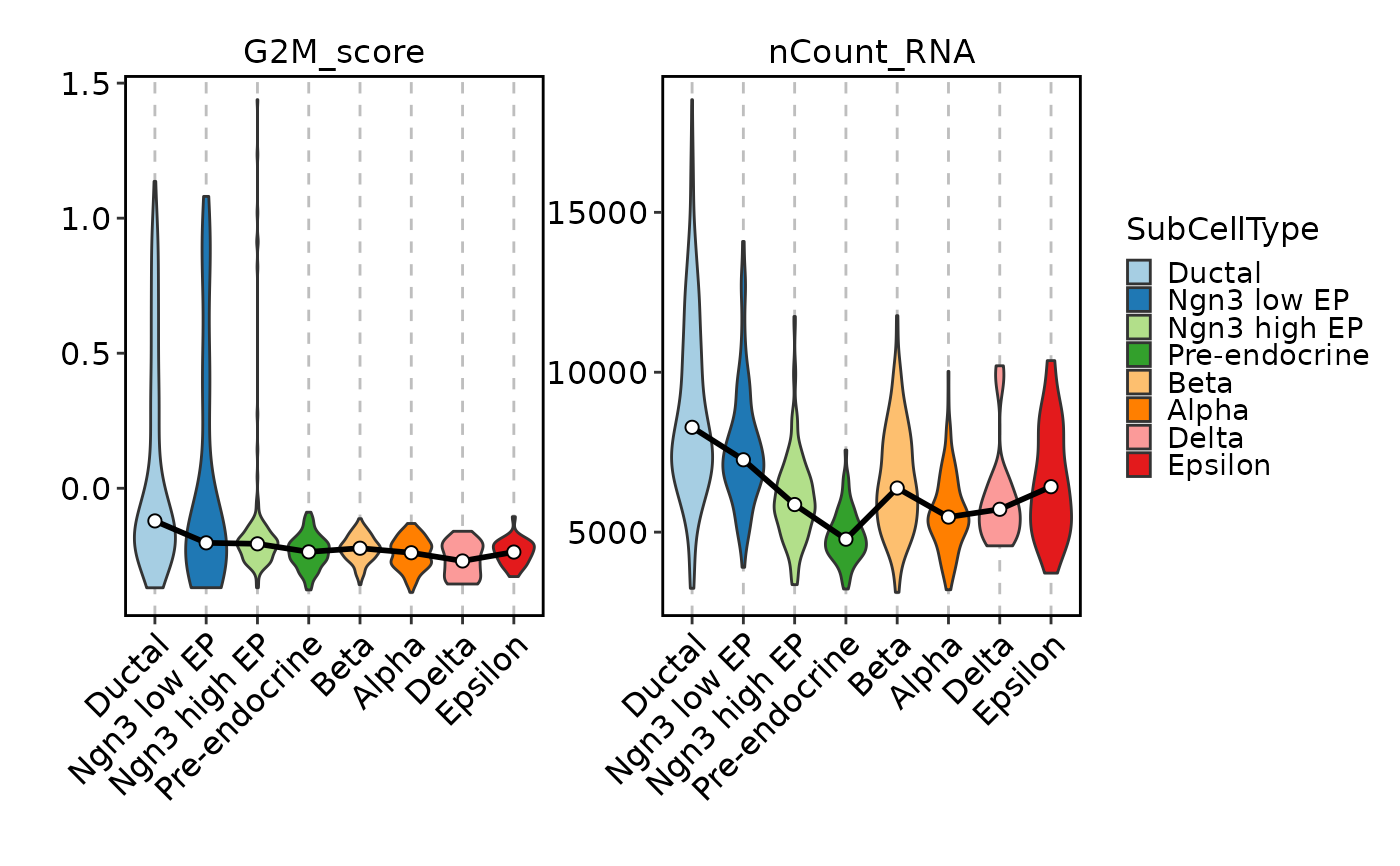
 FeatureStatPlot(pancreas_sub, features = c("G2M_score", "nCount_RNA"),
ident = "SubCellType", facet_scales = "free_y", add_trend = TRUE)
FeatureStatPlot(pancreas_sub, features = c("G2M_score", "nCount_RNA"),
ident = "SubCellType", facet_scales = "free_y", add_trend = TRUE)
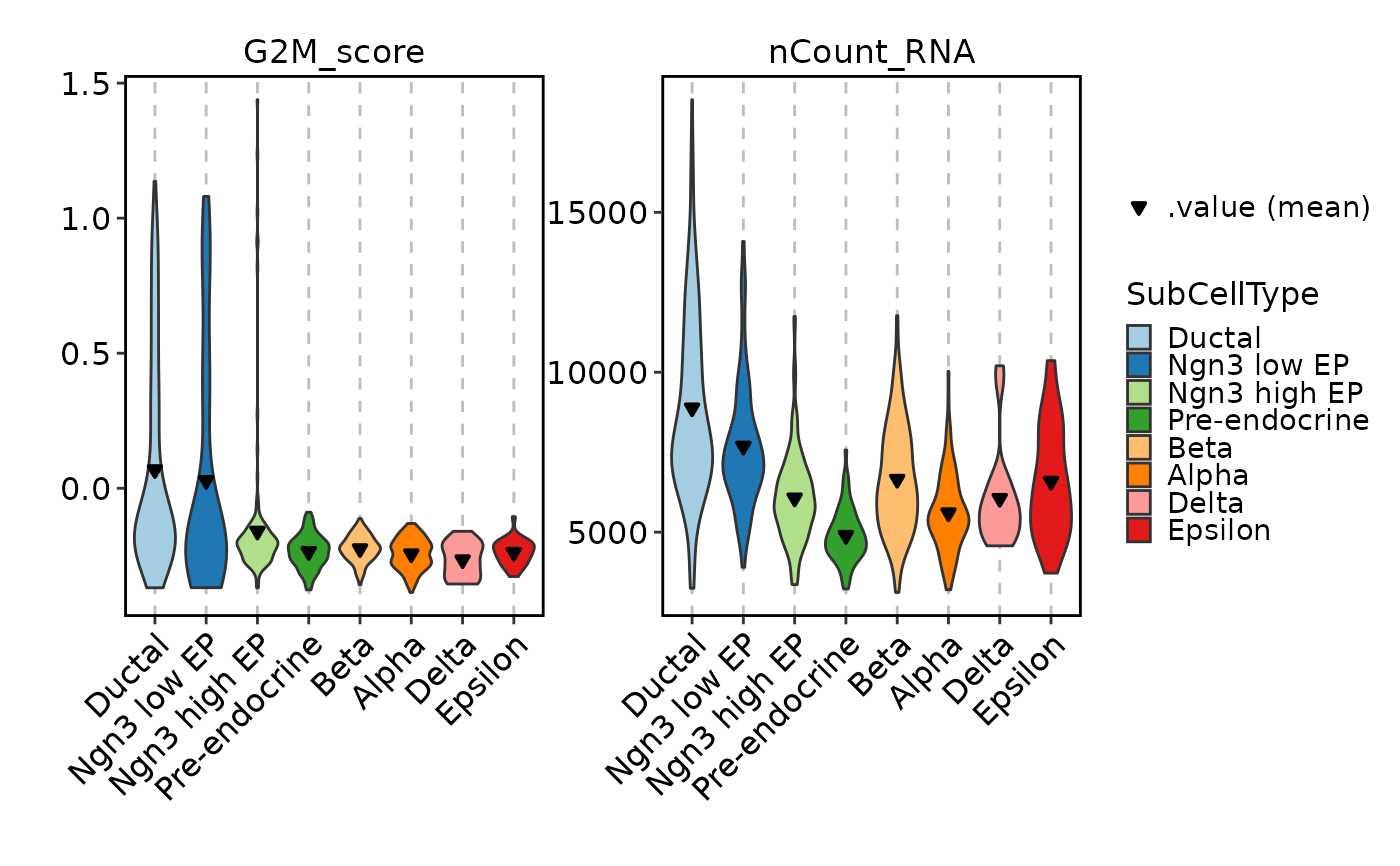
 FeatureStatPlot(pancreas_sub, features = c("G2M_score", "nCount_RNA"),
ident = "SubCellType", facet_scales = "free_y", add_stat = mean)
FeatureStatPlot(pancreas_sub, features = c("G2M_score", "nCount_RNA"),
ident = "SubCellType", facet_scales = "free_y", add_stat = mean)
 FeatureStatPlot(pancreas_sub, features = c("G2M_score", "nCount_RNA"),
ident = "SubCellType", facet_scales = "free_y", group_by = "Phase")
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
FeatureStatPlot(pancreas_sub, features = c("G2M_score", "nCount_RNA"),
ident = "SubCellType", facet_scales = "free_y", group_by = "Phase")
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
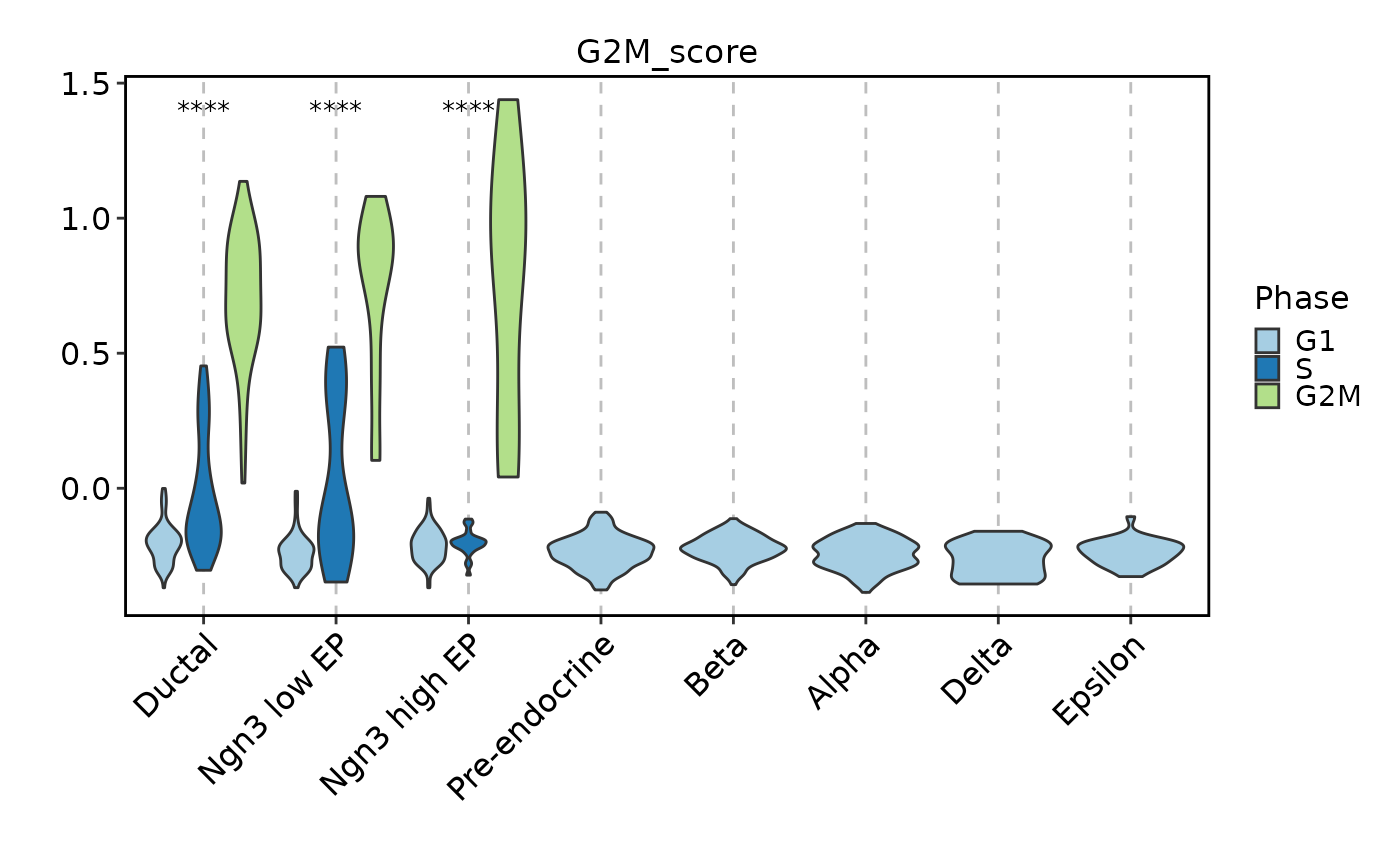
 FeatureStatPlot(
subset(pancreas_sub,
subset = SubCellType %in% c("Ductal", "Ngn3 low EP", "Ngn3 high EP")),
features = c("G2M_score"),
ident = "SubCellType", group_by = "Phase", comparisons = TRUE)
#> Detected more than 2 groups. Use multiple_method for comparison
FeatureStatPlot(
subset(pancreas_sub,
subset = SubCellType %in% c("Ductal", "Ngn3 low EP", "Ngn3 high EP")),
features = c("G2M_score"),
ident = "SubCellType", group_by = "Phase", comparisons = TRUE)
#> Detected more than 2 groups. Use multiple_method for comparison
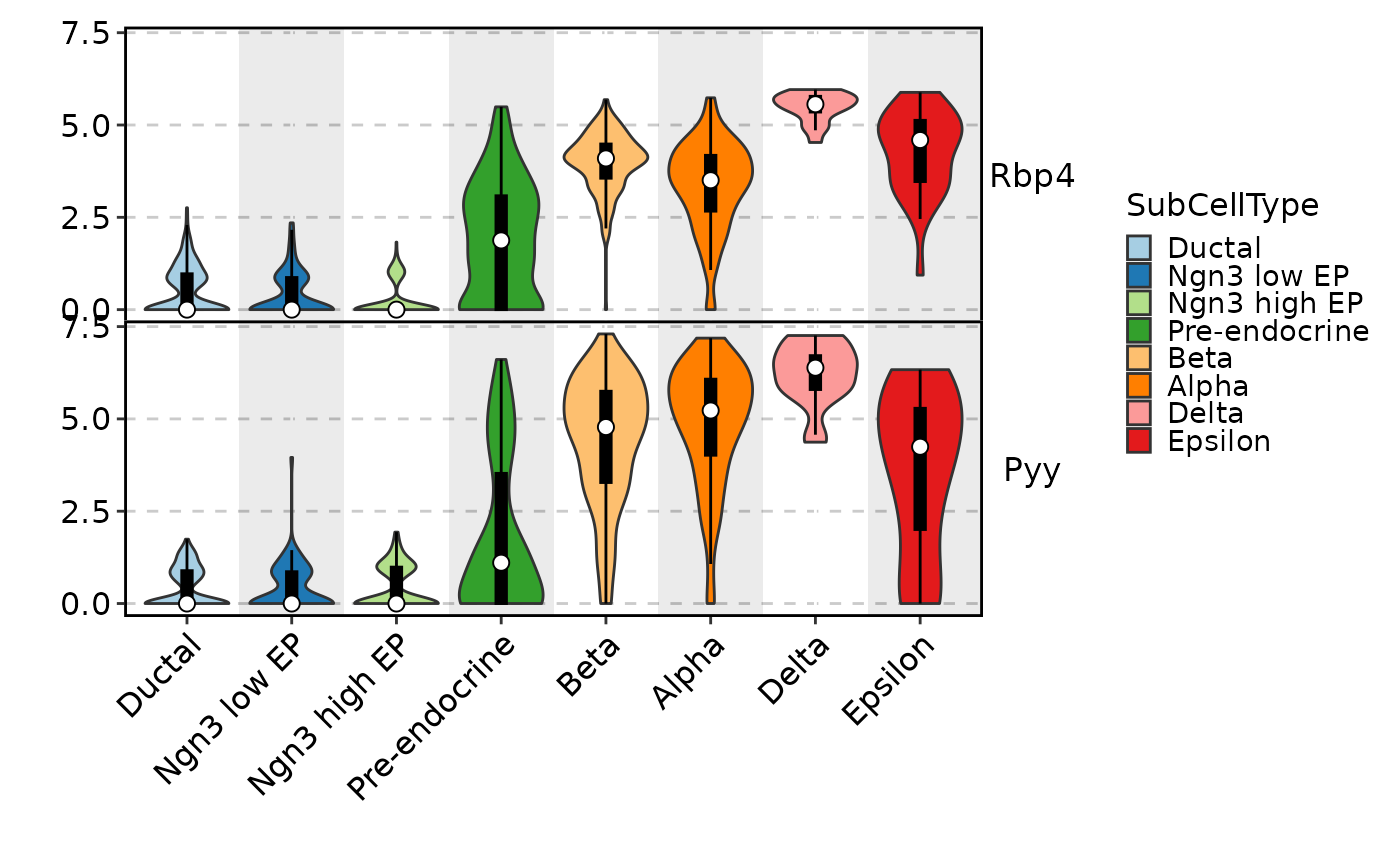
 FeatureStatPlot(pancreas_sub, features = c("Rbp4", "Pyy"), ident = "SubCellType",
add_bg = TRUE, add_box = TRUE, stack = TRUE)
FeatureStatPlot(pancreas_sub, features = c("Rbp4", "Pyy"), ident = "SubCellType",
add_bg = TRUE, add_box = TRUE, stack = TRUE)
 # Use `pos_only` to include only cells with positive expression of all features
FeatureStatPlot(pancreas_sub, features = c("Rbp4", "Pyy"), ident = "SubCellType",
add_bg = TRUE, add_box = TRUE, stack = TRUE, pos_only = "all")
# Use `pos_only` to include only cells with positive expression of all features
FeatureStatPlot(pancreas_sub, features = c("Rbp4", "Pyy"), ident = "SubCellType",
add_bg = TRUE, add_box = TRUE, stack = TRUE, pos_only = "all")
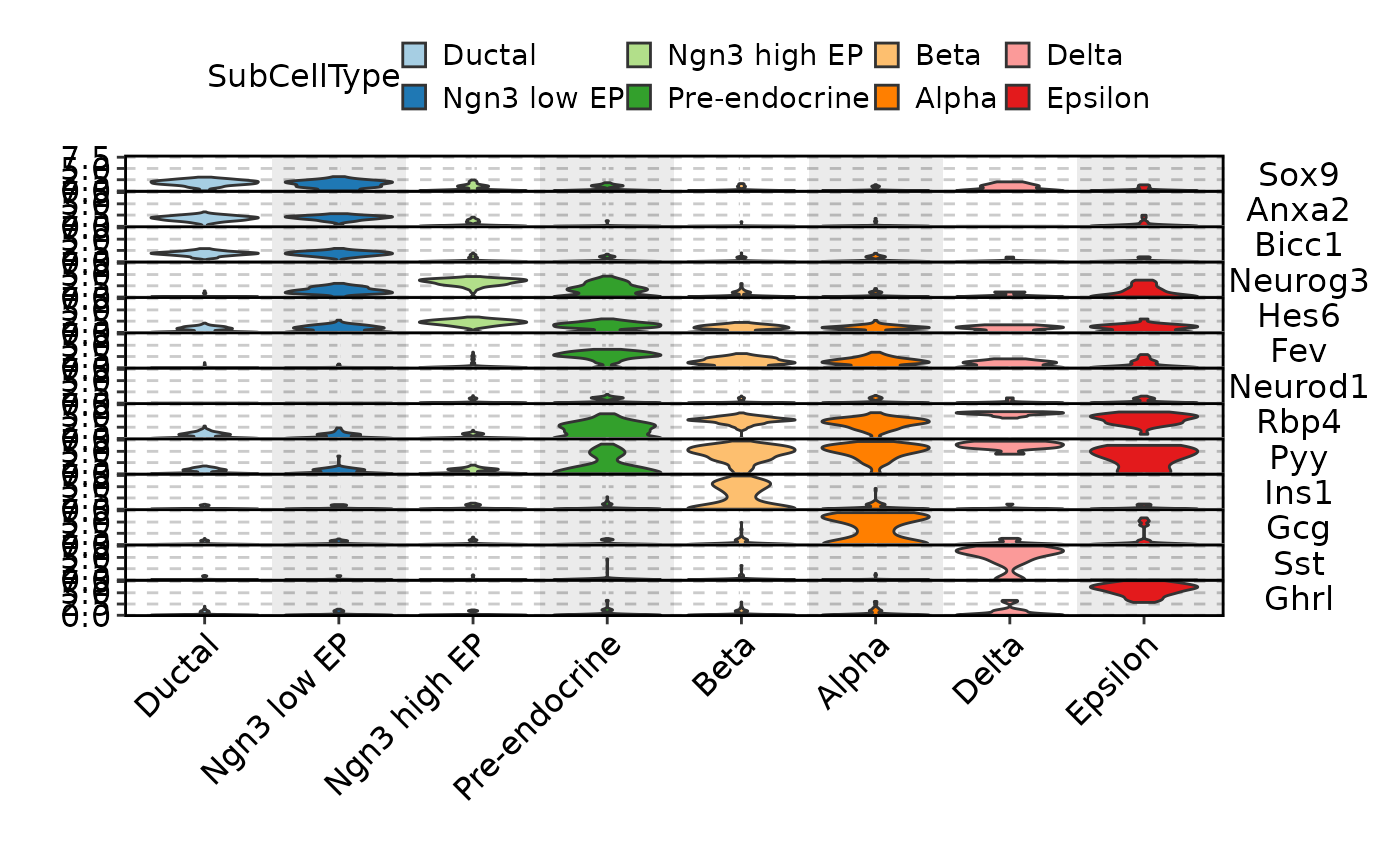 FeatureStatPlot(pancreas_sub, features = c(
"Sox9", "Anxa2", "Bicc1", # Ductal
"Neurog3", "Hes6", # EPs
"Fev", "Neurod1", # Pre-endocrine
"Rbp4", "Pyy", # Endocrine
"Ins1", "Gcg", "Sst", "Ghrl" # Beta, Alpha, Delta, Epsilon
), ident = "SubCellType", add_bg = TRUE, stack = TRUE,
legend.position = "top", legend.direction = "horizontal")
FeatureStatPlot(pancreas_sub, features = c(
"Sox9", "Anxa2", "Bicc1", # Ductal
"Neurog3", "Hes6", # EPs
"Fev", "Neurod1", # Pre-endocrine
"Rbp4", "Pyy", # Endocrine
"Ins1", "Gcg", "Sst", "Ghrl" # Beta, Alpha, Delta, Epsilon
), ident = "SubCellType", add_bg = TRUE, stack = TRUE,
legend.position = "top", legend.direction = "horizontal")
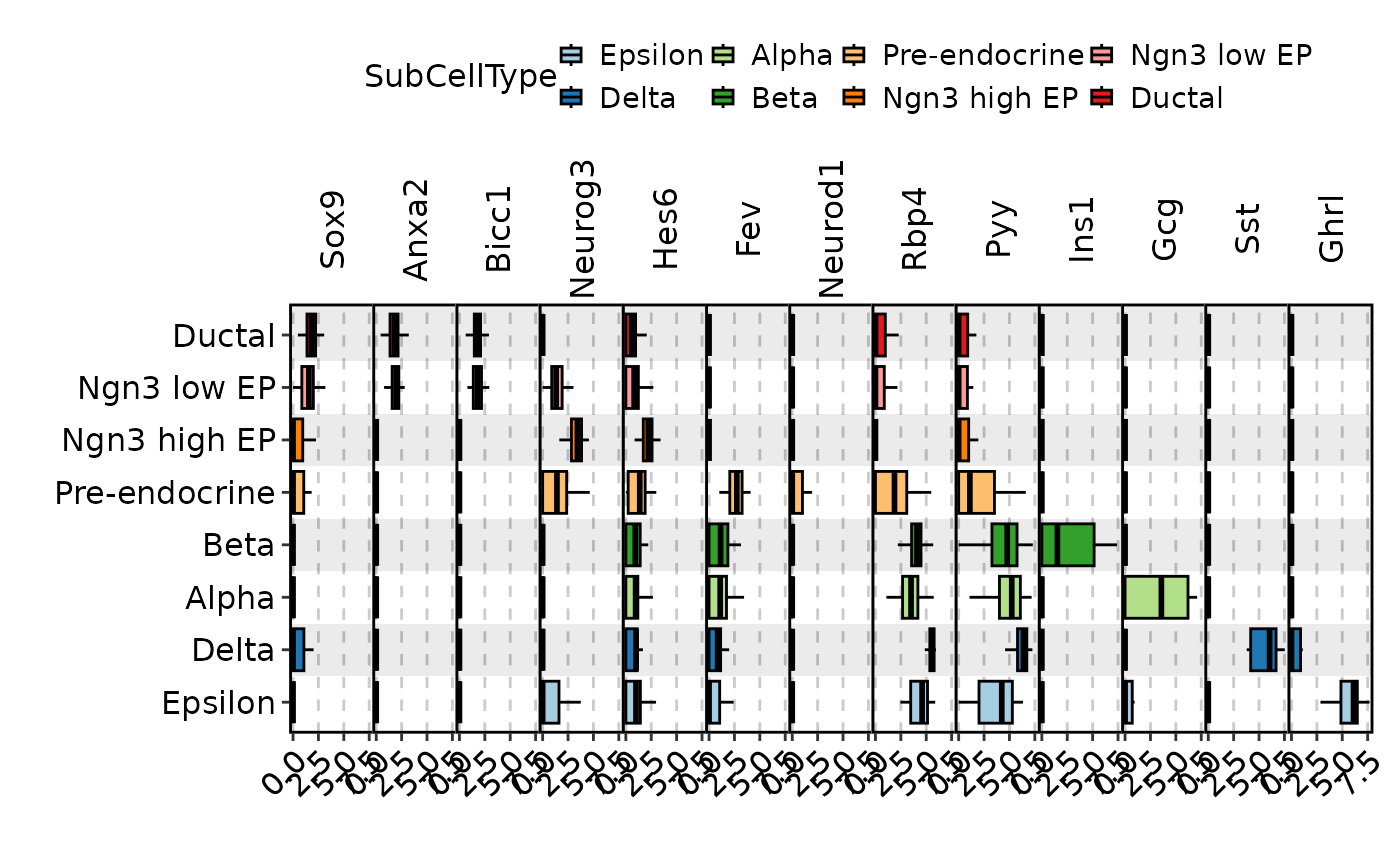 FeatureStatPlot(pancreas_sub, plot_type = "box", features = c(
"Sox9", "Anxa2", "Bicc1", # Ductal
"Neurog3", "Hes6", # EPs
"Fev", "Neurod1", # Pre-endocrine
"Rbp4", "Pyy", # Endocrine
"Ins1", "Gcg", "Sst", "Ghrl" # Beta, Alpha, Delta, Epsilon
), ident = "SubCellType", add_bg = TRUE, stack = TRUE, flip = TRUE,
legend.position = "top", legend.direction = "horizontal")
FeatureStatPlot(pancreas_sub, plot_type = "box", features = c(
"Sox9", "Anxa2", "Bicc1", # Ductal
"Neurog3", "Hes6", # EPs
"Fev", "Neurod1", # Pre-endocrine
"Rbp4", "Pyy", # Endocrine
"Ins1", "Gcg", "Sst", "Ghrl" # Beta, Alpha, Delta, Epsilon
), ident = "SubCellType", add_bg = TRUE, stack = TRUE, flip = TRUE,
legend.position = "top", legend.direction = "horizontal")
 # Use splitting instead of facetting
FeatureStatPlot(pancreas_sub, features = c("Neurog3", "Rbp4", "Ins1"),
ident = "CellType", split_by = TRUE)
# Use splitting instead of facetting
FeatureStatPlot(pancreas_sub, features = c("Neurog3", "Rbp4", "Ins1"),
ident = "CellType", split_by = TRUE)
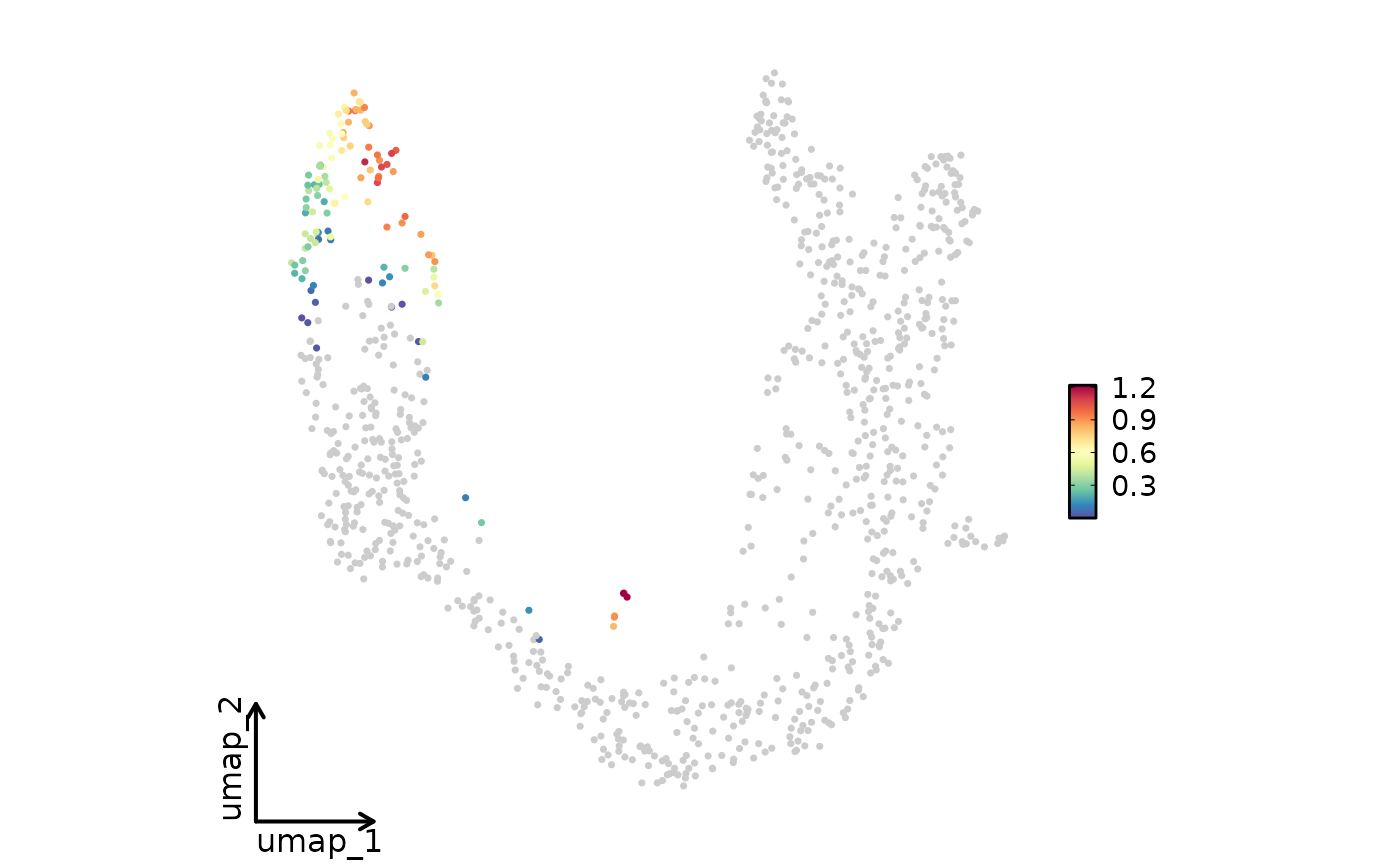
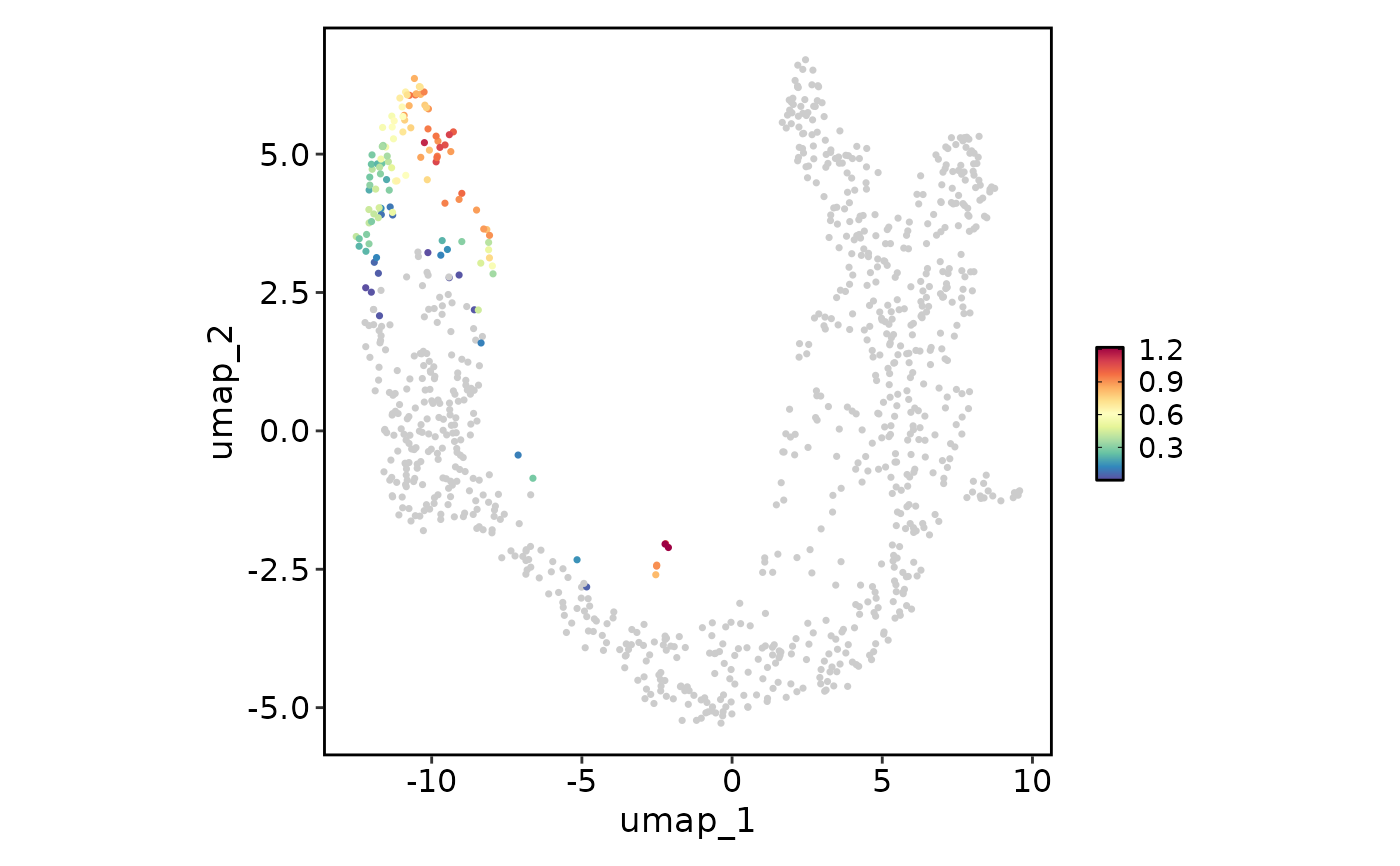 FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "G2M_score", reduction = "UMAP")
FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "G2M_score", reduction = "UMAP")
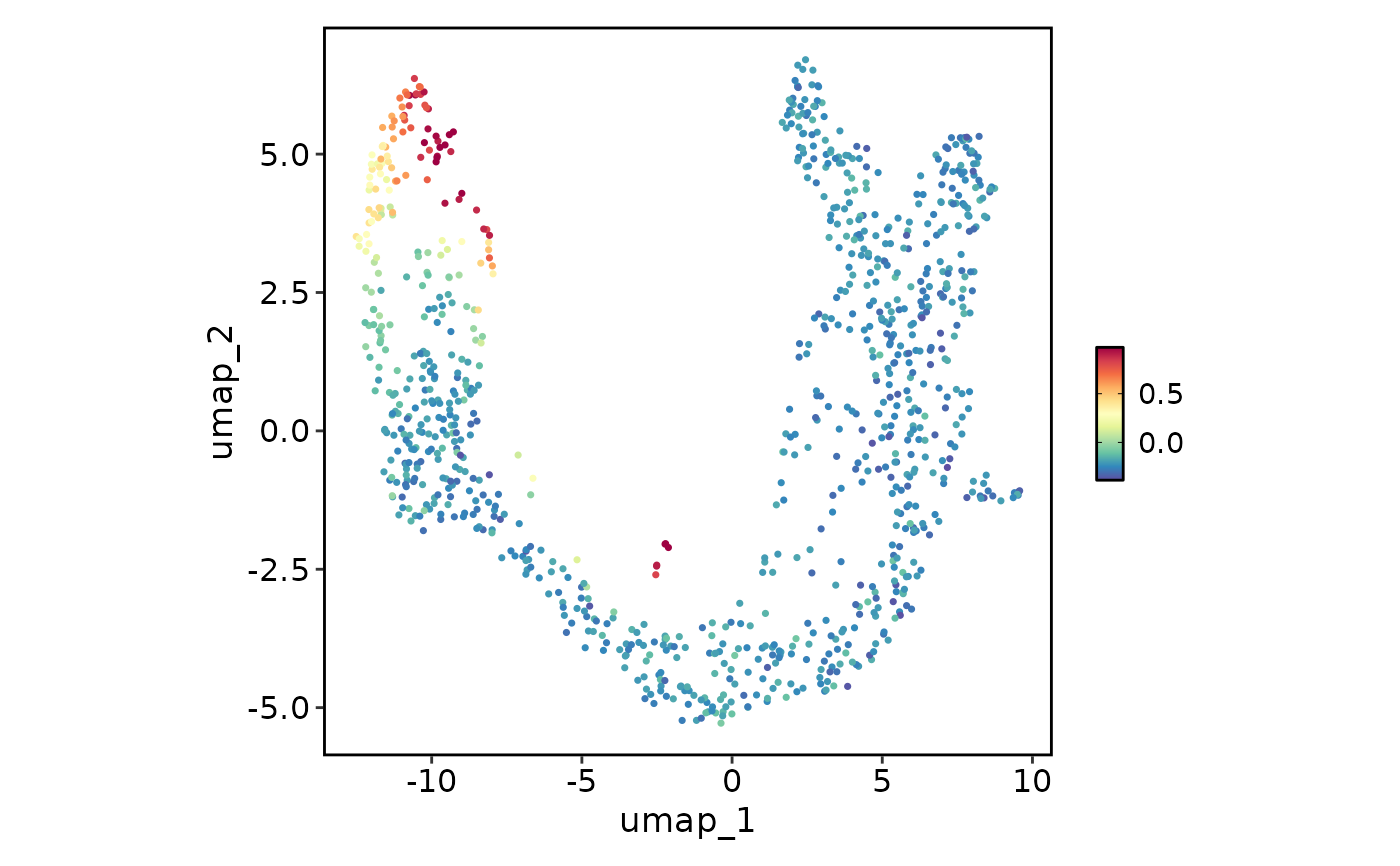 FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "G2M_score", reduction = "UMAP",
bg_cutoff = -Inf)
FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "G2M_score", reduction = "UMAP",
bg_cutoff = -Inf)
 FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "G2M_score", reduction = "UMAP",
theme = "theme_blank")
FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "G2M_score", reduction = "UMAP",
theme = "theme_blank")
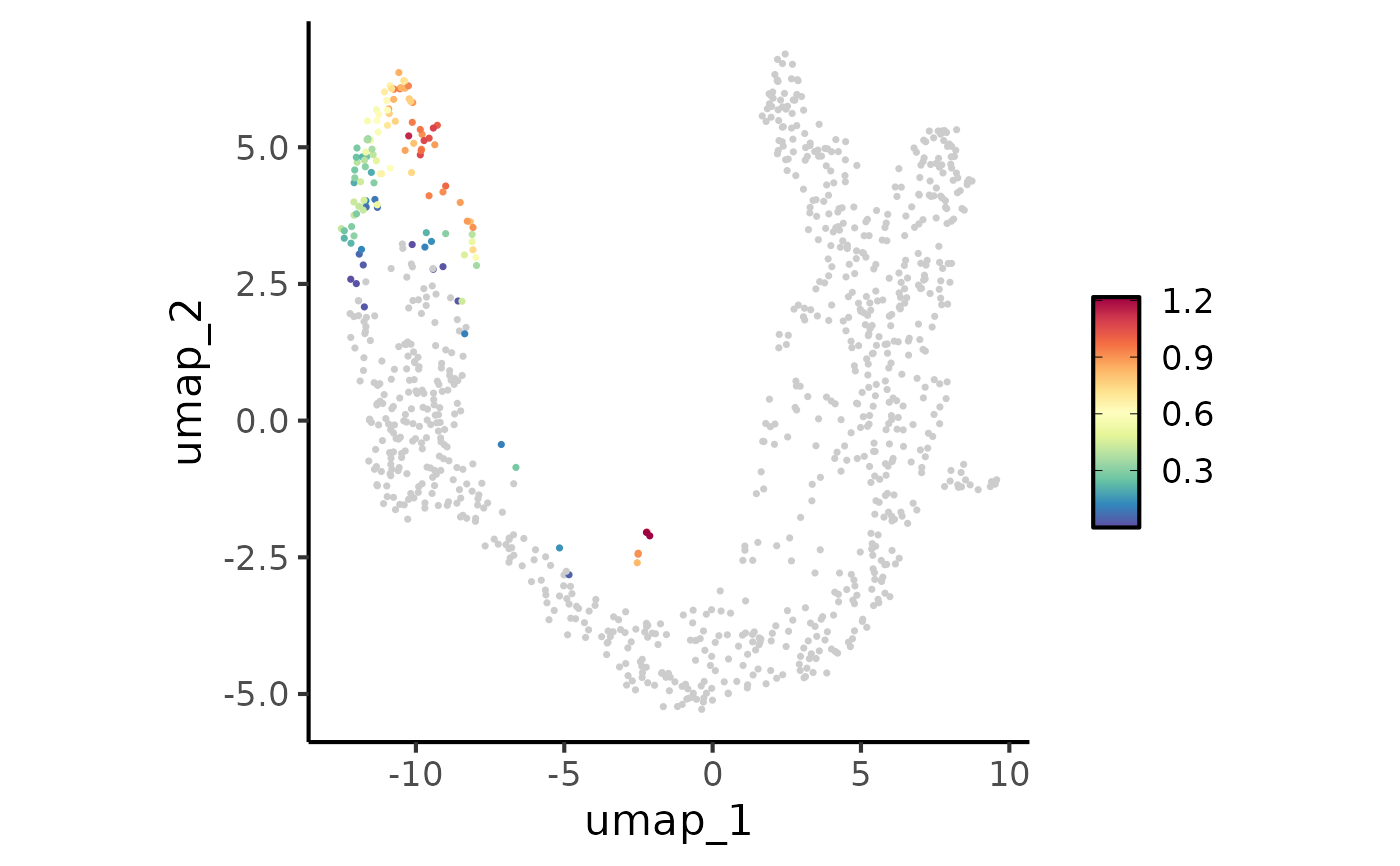 FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "G2M_score", reduction = "UMAP",
theme = ggplot2::theme_classic, theme_args = list(base_size = 16))
FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "G2M_score", reduction = "UMAP",
theme = ggplot2::theme_classic, theme_args = list(base_size = 16))
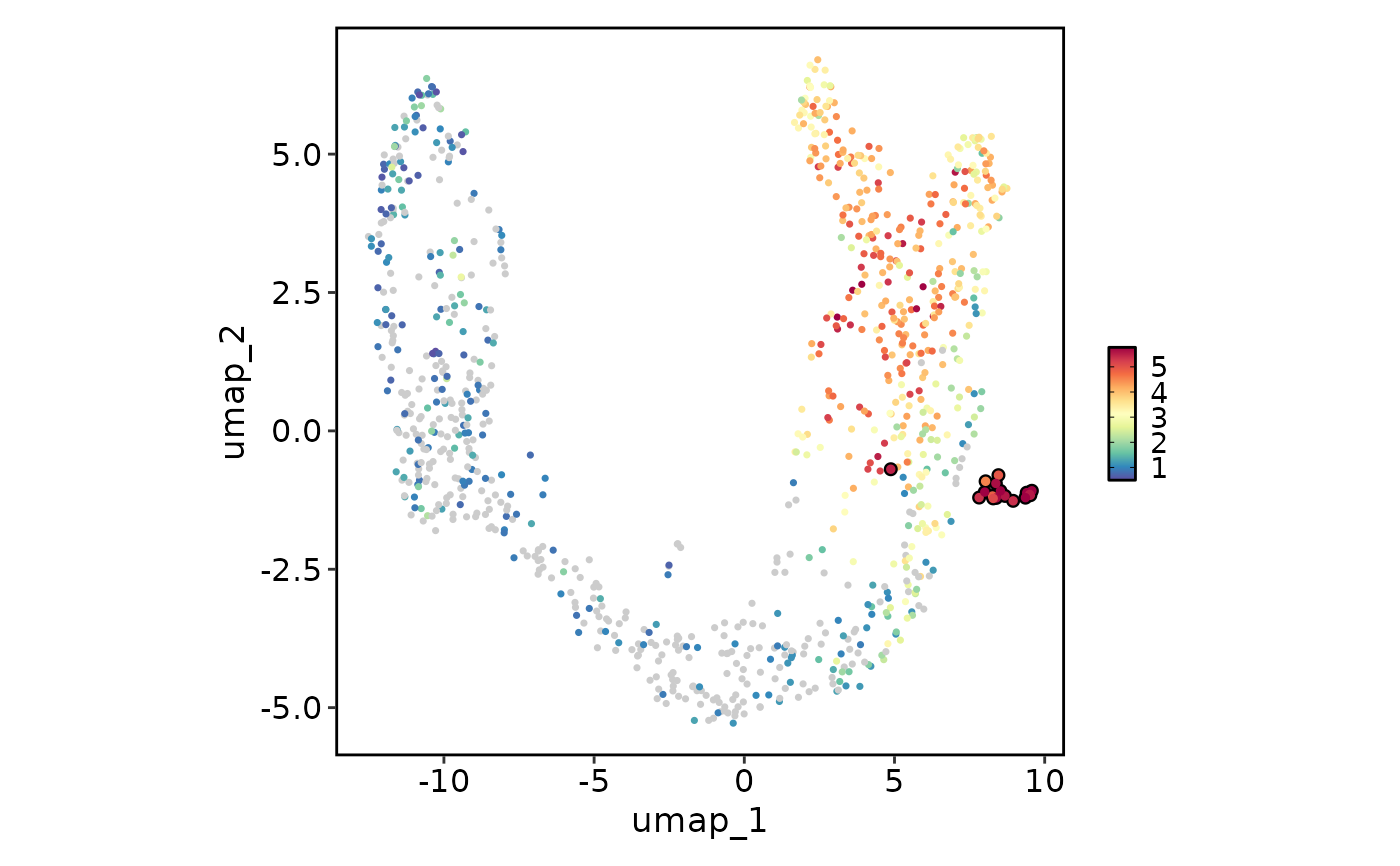 # Label and highlight cell points
FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "Rbp4", reduction = "UMAP",
highlight = 'SubCellType == "Delta"')
# Label and highlight cell points
FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "Rbp4", reduction = "UMAP",
highlight = 'SubCellType == "Delta"')
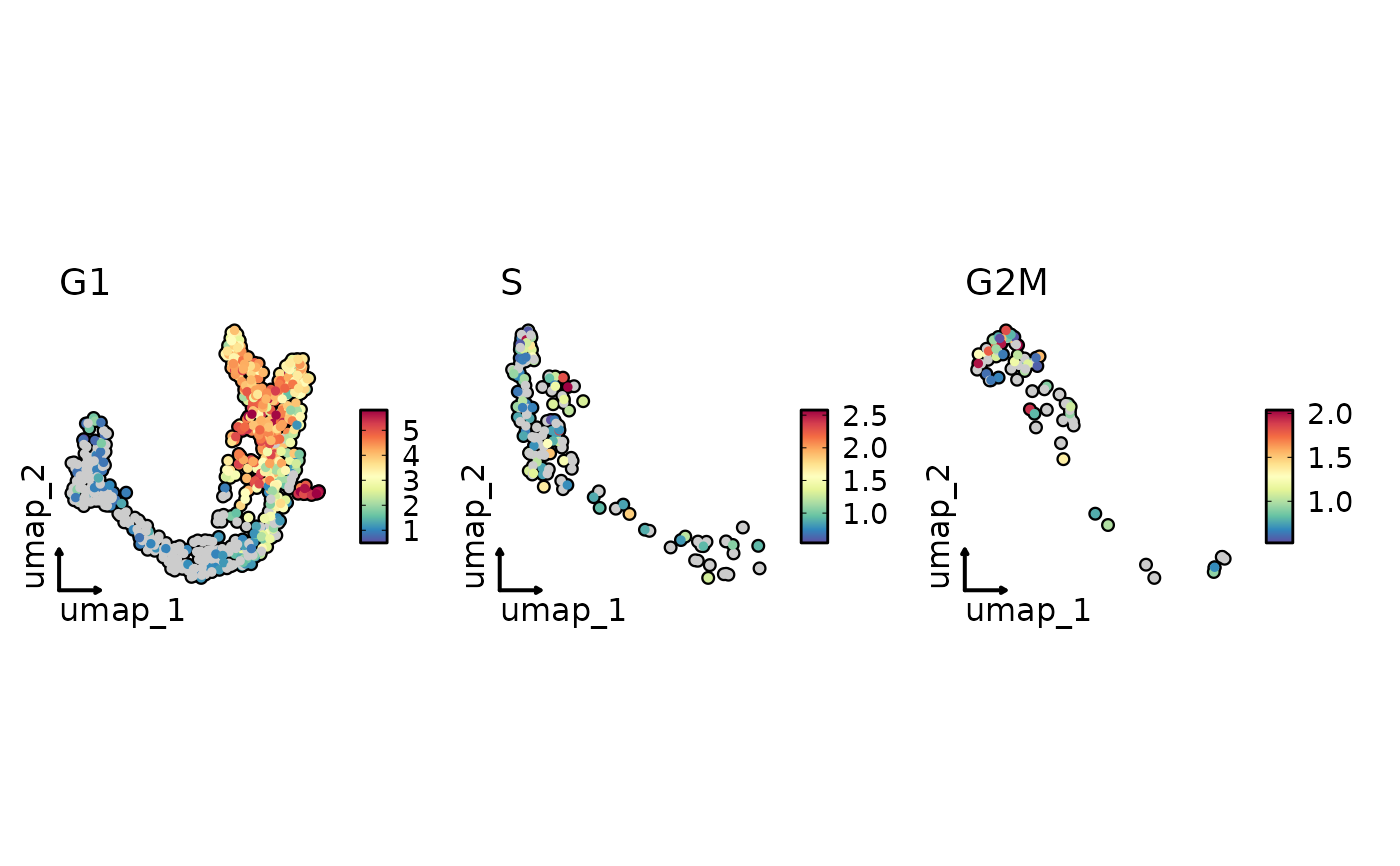 FeatureStatPlot(pancreas_sub, plot_type = "dim",
features = "Rbp4", split_by = "Phase", reduction = "UMAP",
highlight = TRUE, theme = "theme_blank")
FeatureStatPlot(pancreas_sub, plot_type = "dim",
features = "Rbp4", split_by = "Phase", reduction = "UMAP",
highlight = TRUE, theme = "theme_blank")
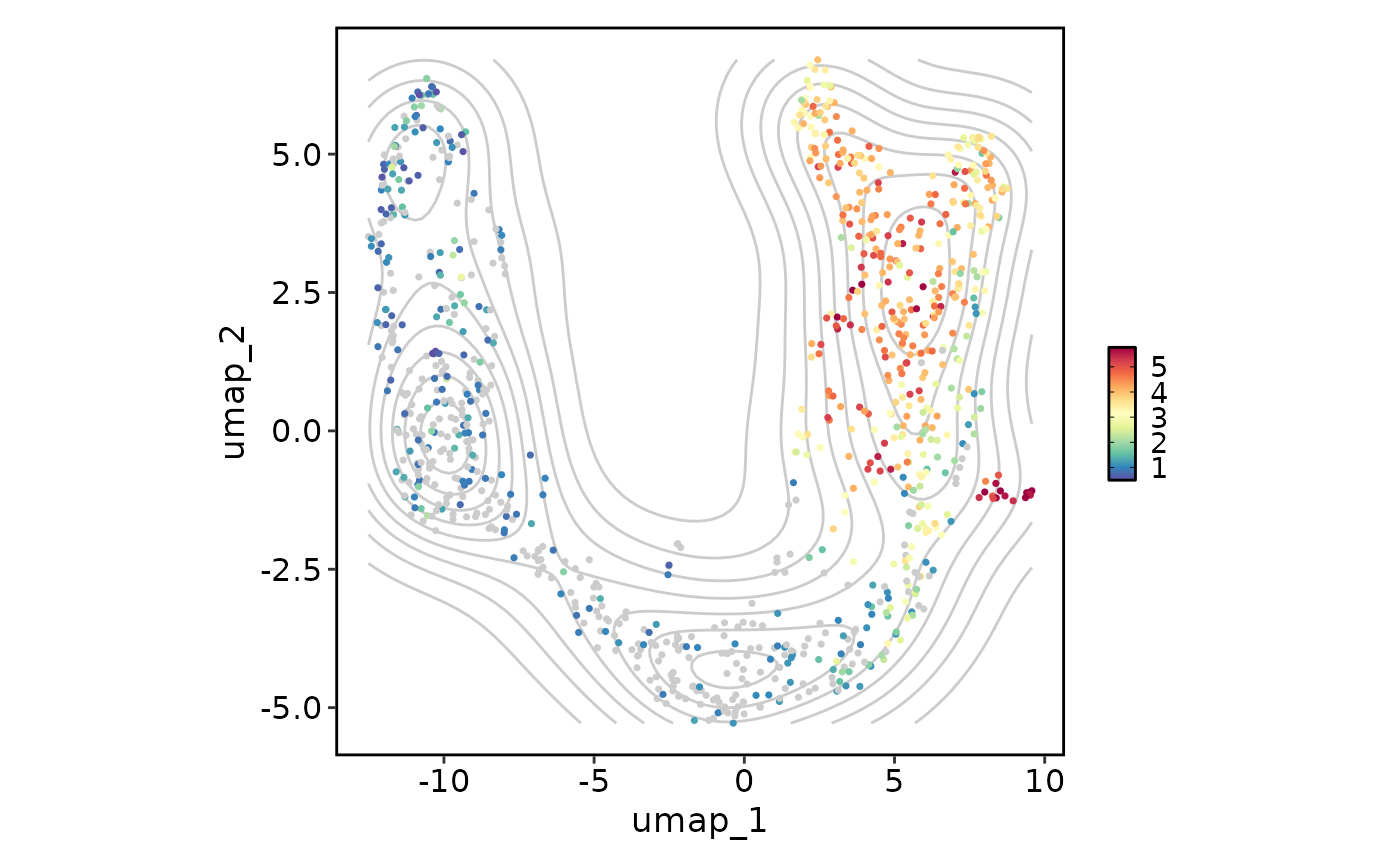 # Add a density layer
FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "Rbp4", reduction = "UMAP",
add_density = TRUE)
# Add a density layer
FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "Rbp4", reduction = "UMAP",
add_density = TRUE)
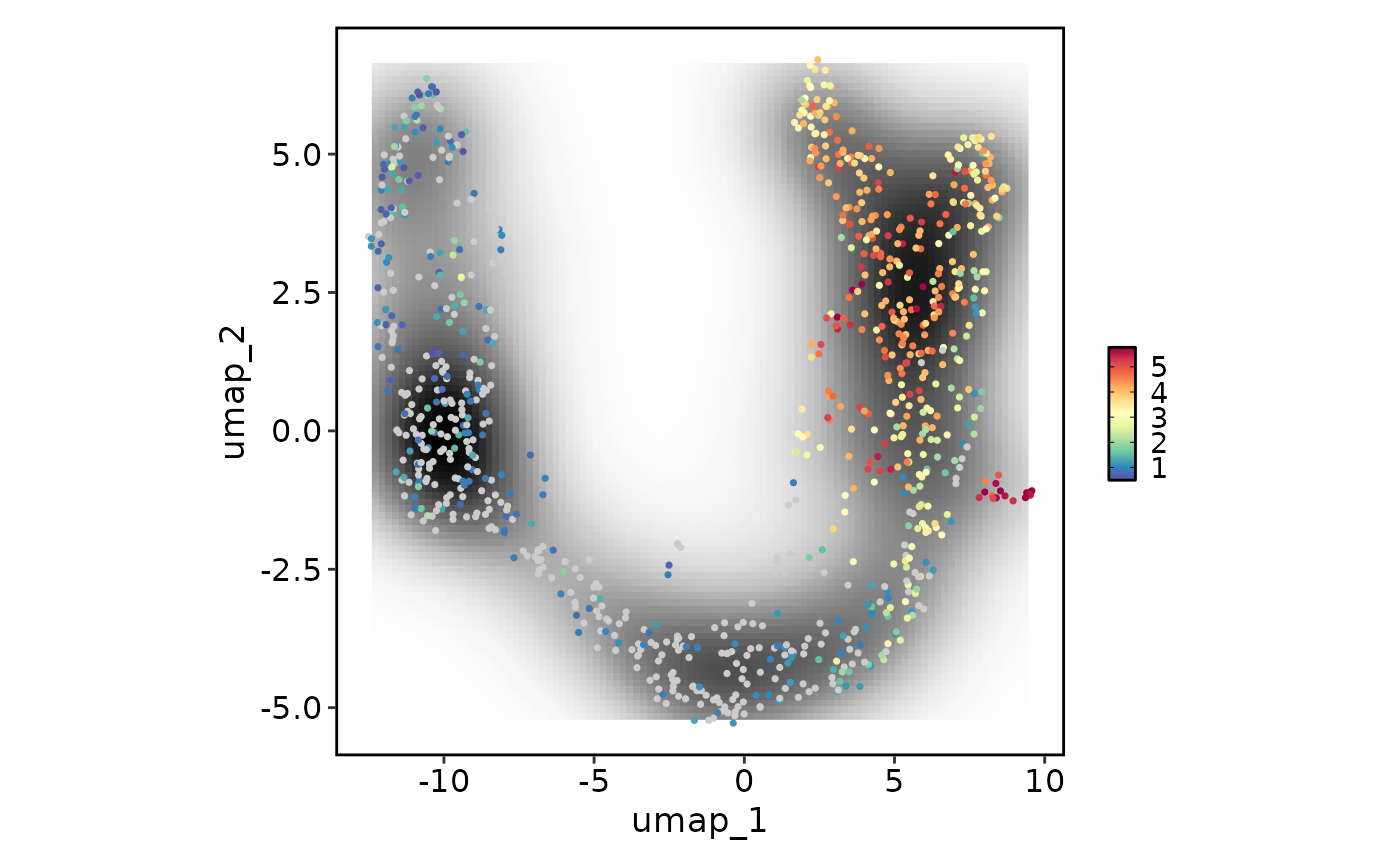 FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "Rbp4", reduction = "UMAP",
add_density = TRUE, density_filled = TRUE)
#> Warning: Removed 396 rows containing missing values or values outside the scale range
#> (`geom_raster()`).
FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "Rbp4", reduction = "UMAP",
add_density = TRUE, density_filled = TRUE)
#> Warning: Removed 396 rows containing missing values or values outside the scale range
#> (`geom_raster()`).
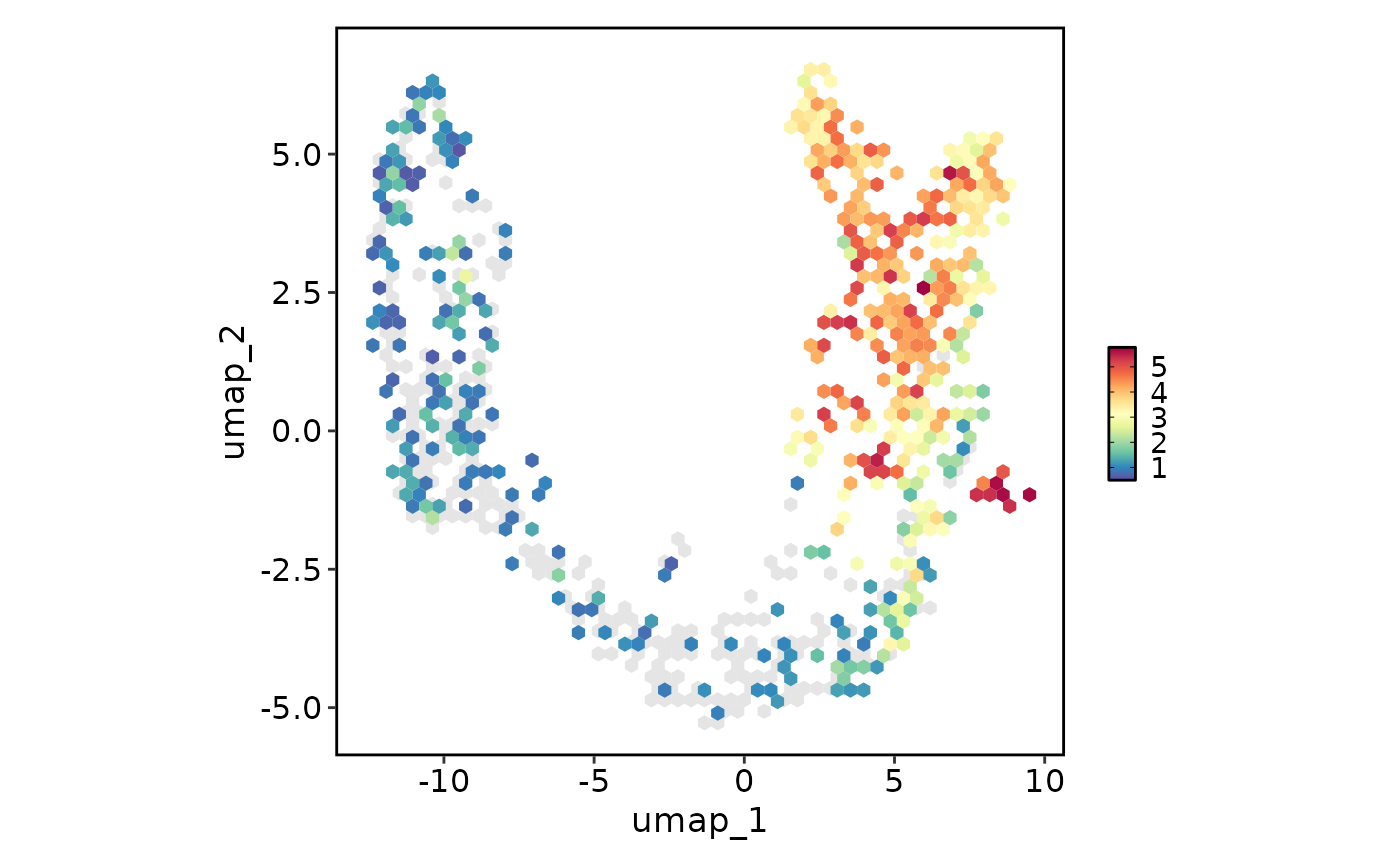 # Change the plot type from point to the hexagonal bin
FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "Rbp4", reduction = "UMAP",
hex = TRUE)
#> Warning: Removed 4 rows containing missing values or values outside the scale range
#> (`geom_hex()`).
# Change the plot type from point to the hexagonal bin
FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "Rbp4", reduction = "UMAP",
hex = TRUE)
#> Warning: Removed 4 rows containing missing values or values outside the scale range
#> (`geom_hex()`).
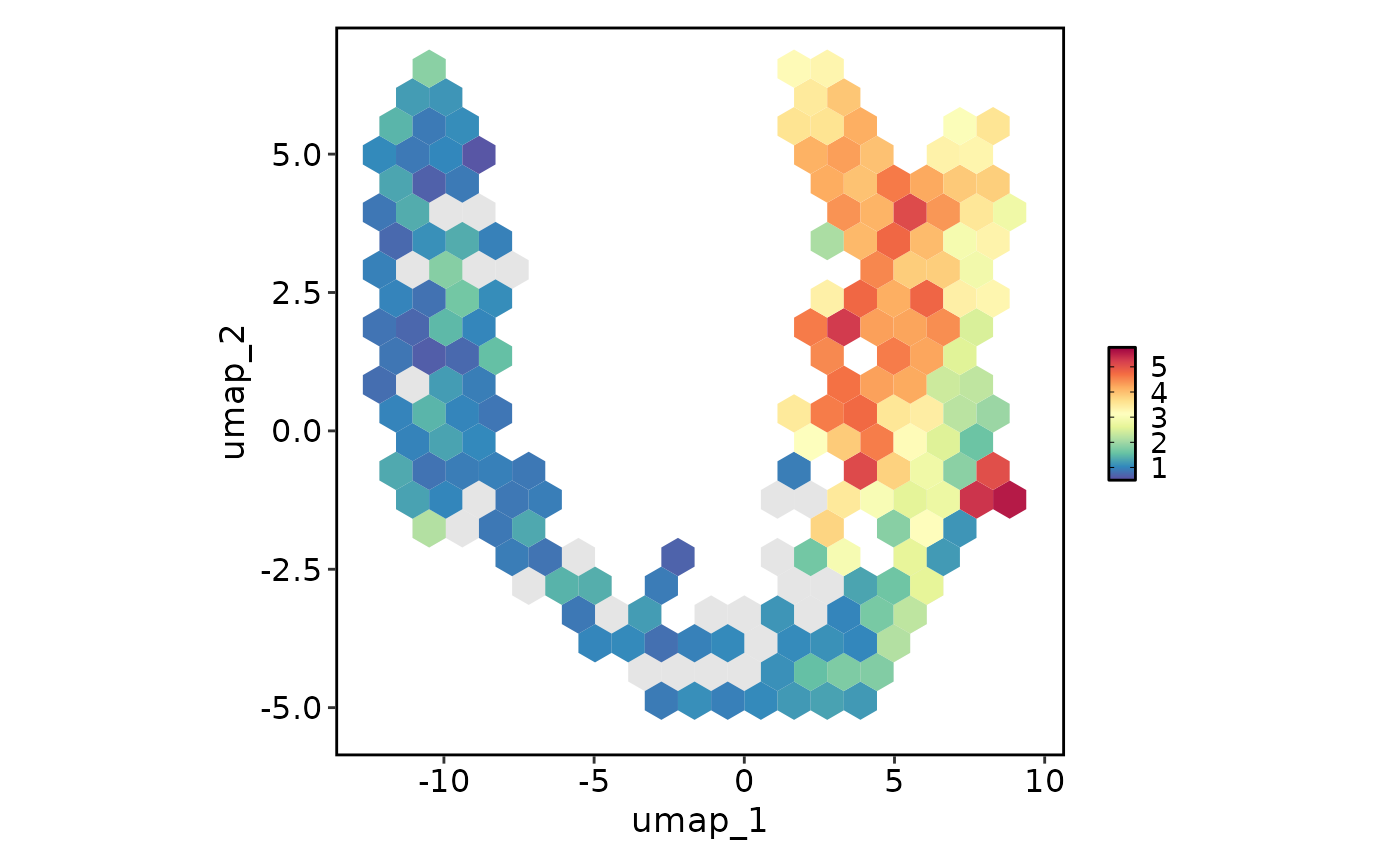 FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "Rbp4", reduction = "UMAP",
hex = TRUE, hex_bins = 20)
#> Warning: Removed 3 rows containing missing values or values outside the scale range
#> (`geom_hex()`).
#> Warning: Removed 5 rows containing missing values or values outside the scale range
#> (`geom_hex()`).
FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "Rbp4", reduction = "UMAP",
hex = TRUE, hex_bins = 20)
#> Warning: Removed 3 rows containing missing values or values outside the scale range
#> (`geom_hex()`).
#> Warning: Removed 5 rows containing missing values or values outside the scale range
#> (`geom_hex()`).
 # Show lineages on the plot based on the pseudotime
FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "Lineage3", reduction = "UMAP",
lineages = "Lineage3")
# Show lineages on the plot based on the pseudotime
FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "Lineage3", reduction = "UMAP",
lineages = "Lineage3")
 FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "Lineage3", reduction = "UMAP",
lineages = "Lineage3", lineages_whiskers = TRUE)
FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "Lineage3", reduction = "UMAP",
lineages = "Lineage3", lineages_whiskers = TRUE)
 FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "Lineage3", reduction = "UMAP",
lineages = "Lineage3", lineages_span = 0.1)
FeatureStatPlot(pancreas_sub, plot_type = "dim", features = "Lineage3", reduction = "UMAP",
lineages = "Lineage3", lineages_span = 0.1)
 FeatureStatPlot(pancreas_sub, plot_type = "dim",
features = c("Sox9", "Anxa2", "Bicc1"), reduction = "UMAP",
theme = "theme_blank",
theme_args = list(plot.subtitle = ggplot2::element_text(size = 10),
strip.text = ggplot2::element_text(size = 8))
)
FeatureStatPlot(pancreas_sub, plot_type = "dim",
features = c("Sox9", "Anxa2", "Bicc1"), reduction = "UMAP",
theme = "theme_blank",
theme_args = list(plot.subtitle = ggplot2::element_text(size = 10),
strip.text = ggplot2::element_text(size = 8))
)
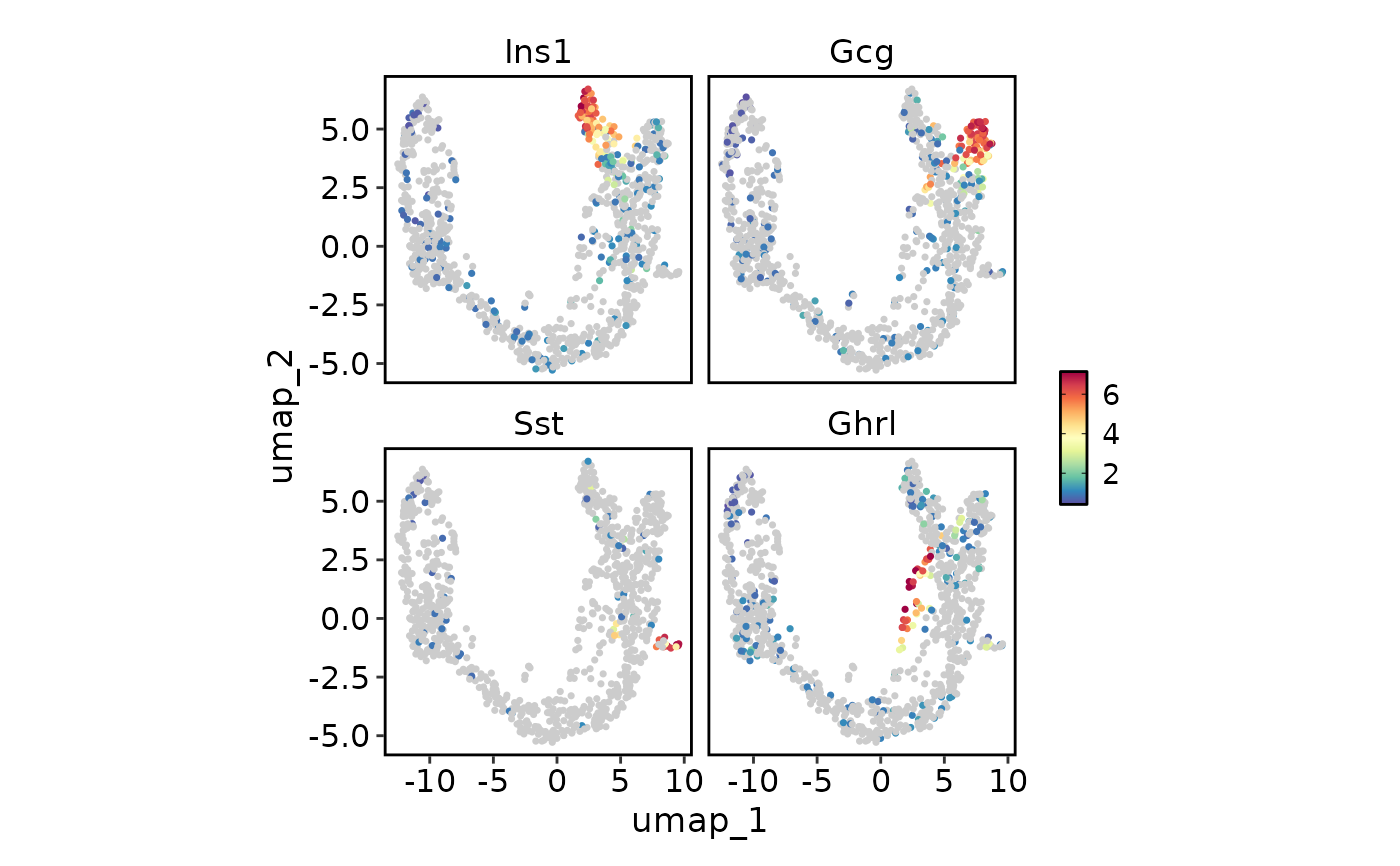 # Plot multiple features with different scales
endocrine_markers <- c("Ins1", "Gcg", "Sst", "Ghrl")
FeatureStatPlot(pancreas_sub, endocrine_markers, reduction = "UMAP", plot_type = "dim")
# Plot multiple features with different scales
endocrine_markers <- c("Ins1", "Gcg", "Sst", "Ghrl")
FeatureStatPlot(pancreas_sub, endocrine_markers, reduction = "UMAP", plot_type = "dim")
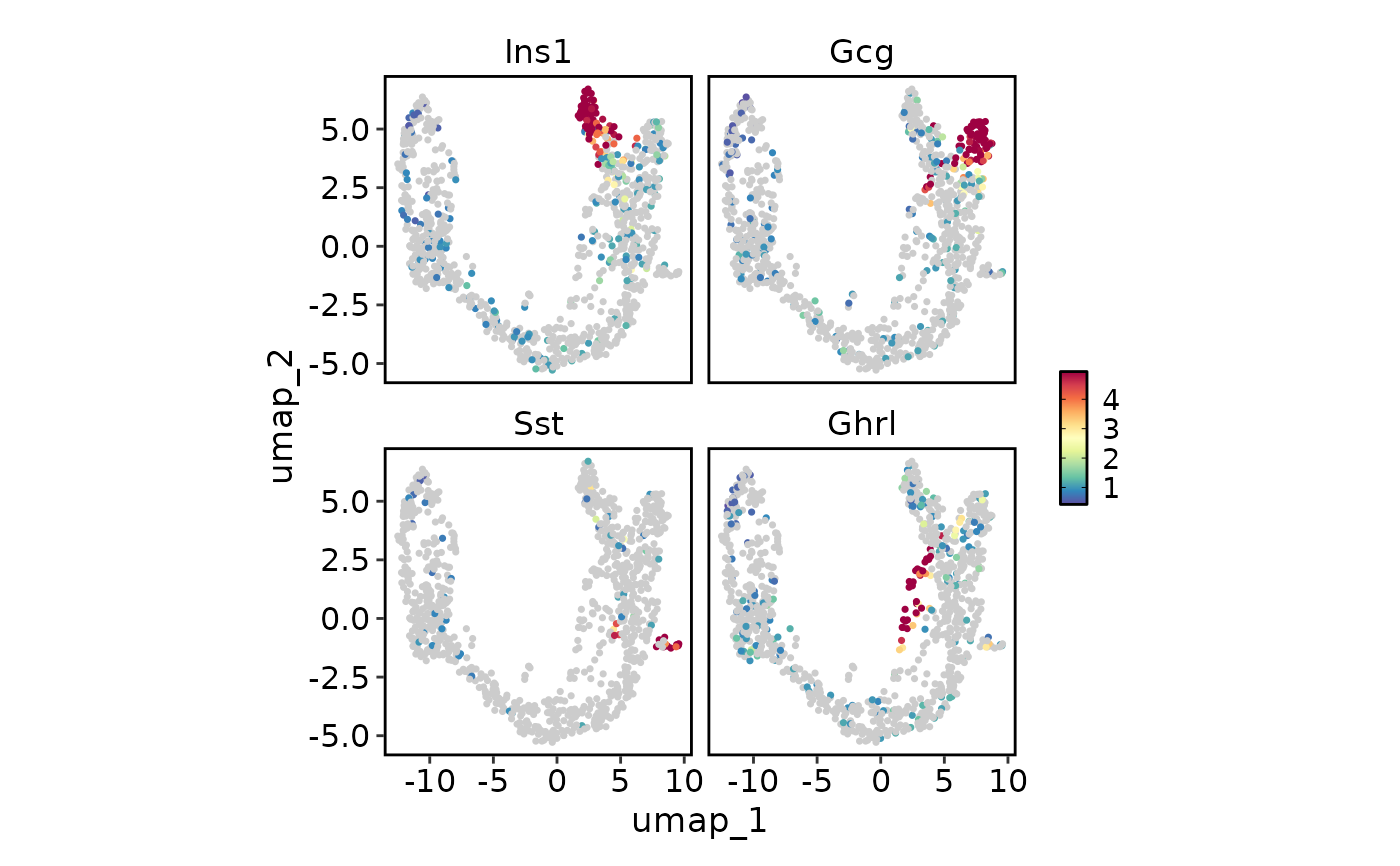 FeatureStatPlot(pancreas_sub, endocrine_markers, reduction = "UMAP", lower_quantile = 0,
upper_quantile = 0.8, plot_type = "dim")
FeatureStatPlot(pancreas_sub, endocrine_markers, reduction = "UMAP", lower_quantile = 0,
upper_quantile = 0.8, plot_type = "dim")
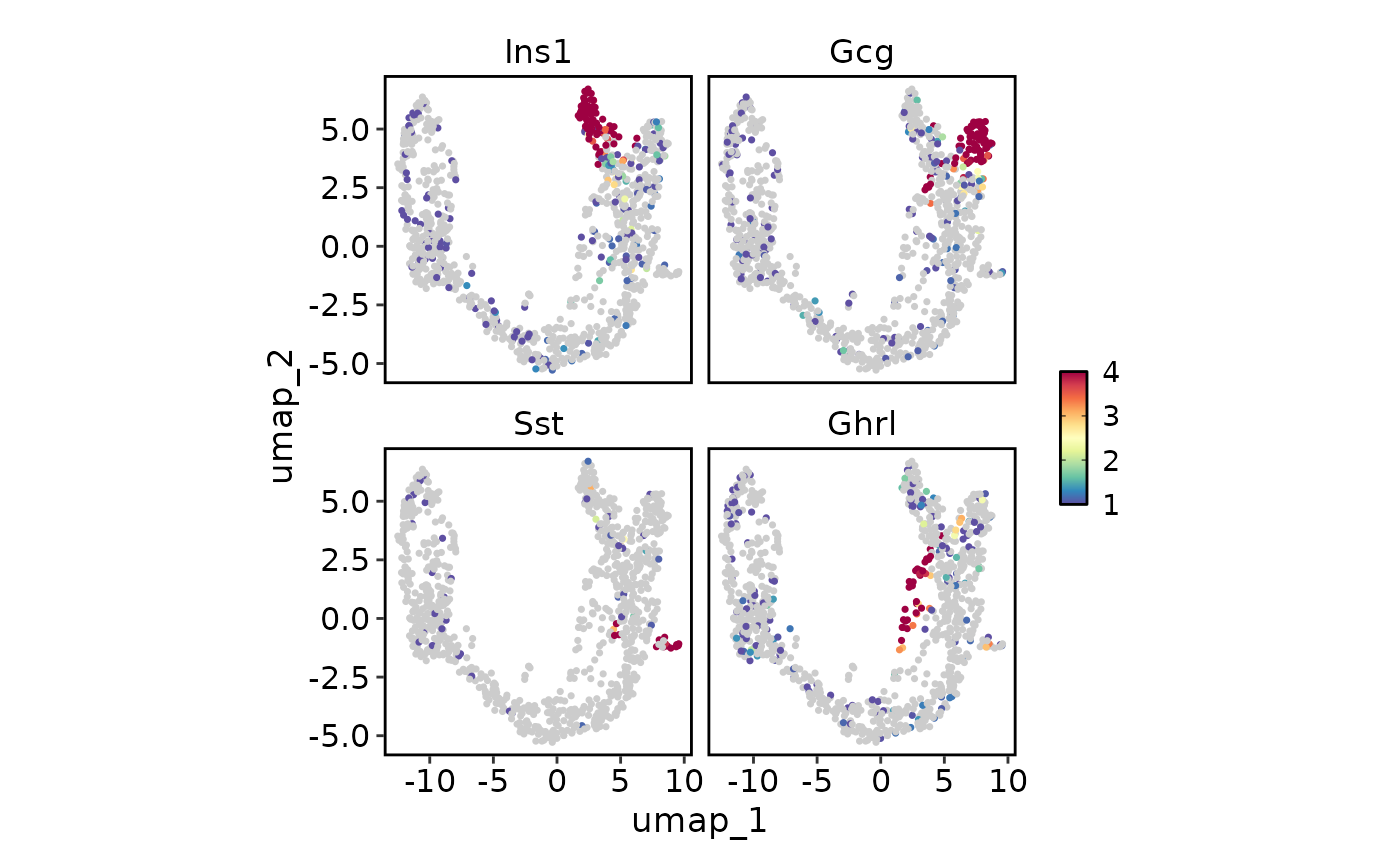 FeatureStatPlot(pancreas_sub, endocrine_markers, reduction = "UMAP",
lower_cutoff = 1, upper_cutoff = 4, plot_type = "dim")
FeatureStatPlot(pancreas_sub, endocrine_markers, reduction = "UMAP",
lower_cutoff = 1, upper_cutoff = 4, plot_type = "dim")
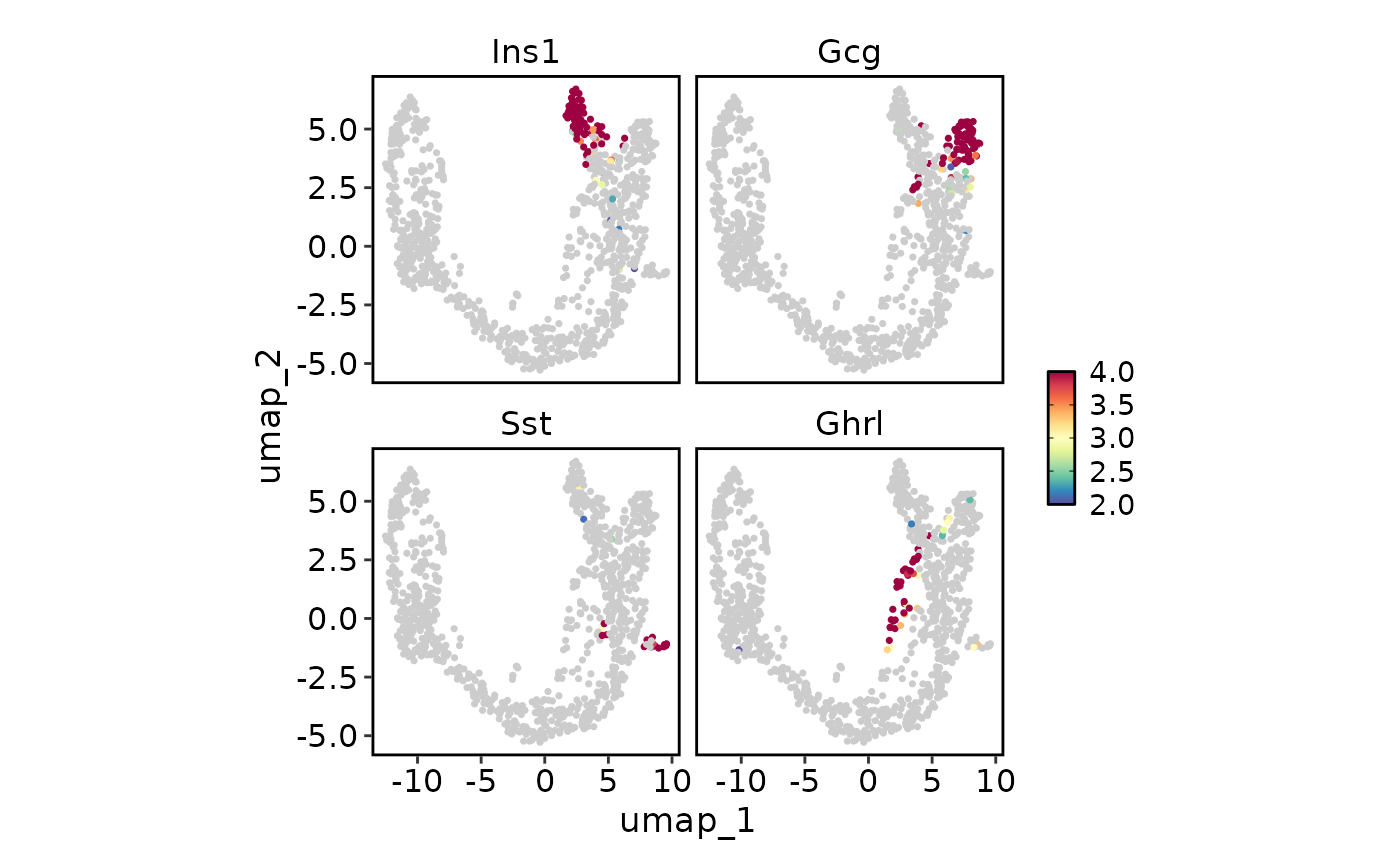 FeatureStatPlot(pancreas_sub, endocrine_markers, reduction = "UMAP", bg_cutoff = 2,
lower_cutoff = 2, upper_cutoff = 4, plot_type = "dim")
FeatureStatPlot(pancreas_sub, endocrine_markers, reduction = "UMAP", bg_cutoff = 2,
lower_cutoff = 2, upper_cutoff = 4, plot_type = "dim")
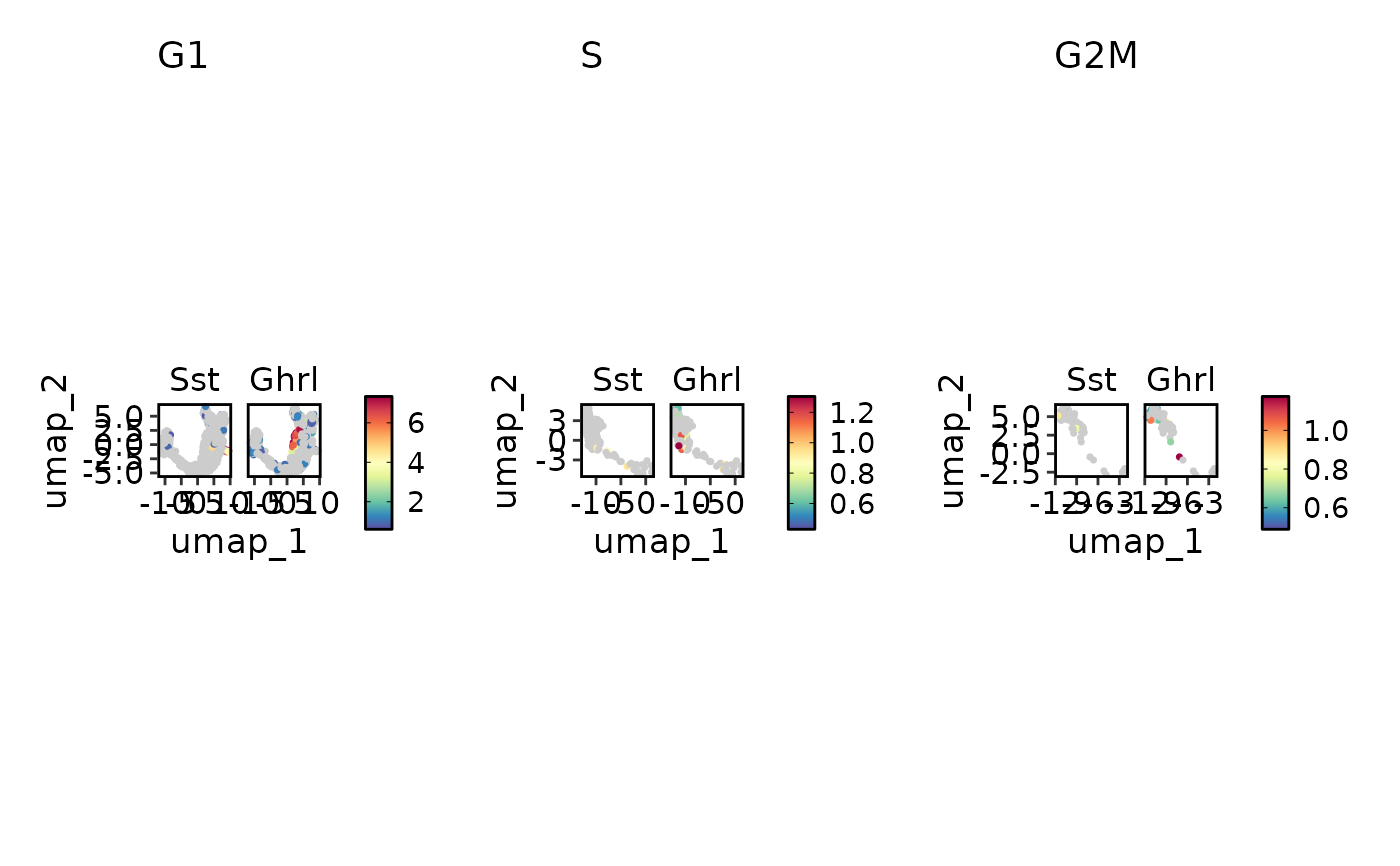 FeatureStatPlot(pancreas_sub, c("Sst", "Ghrl"), split_by = "Phase", reduction = "UMAP",
plot_type = "dim")
FeatureStatPlot(pancreas_sub, c("Sst", "Ghrl"), split_by = "Phase", reduction = "UMAP",
plot_type = "dim")
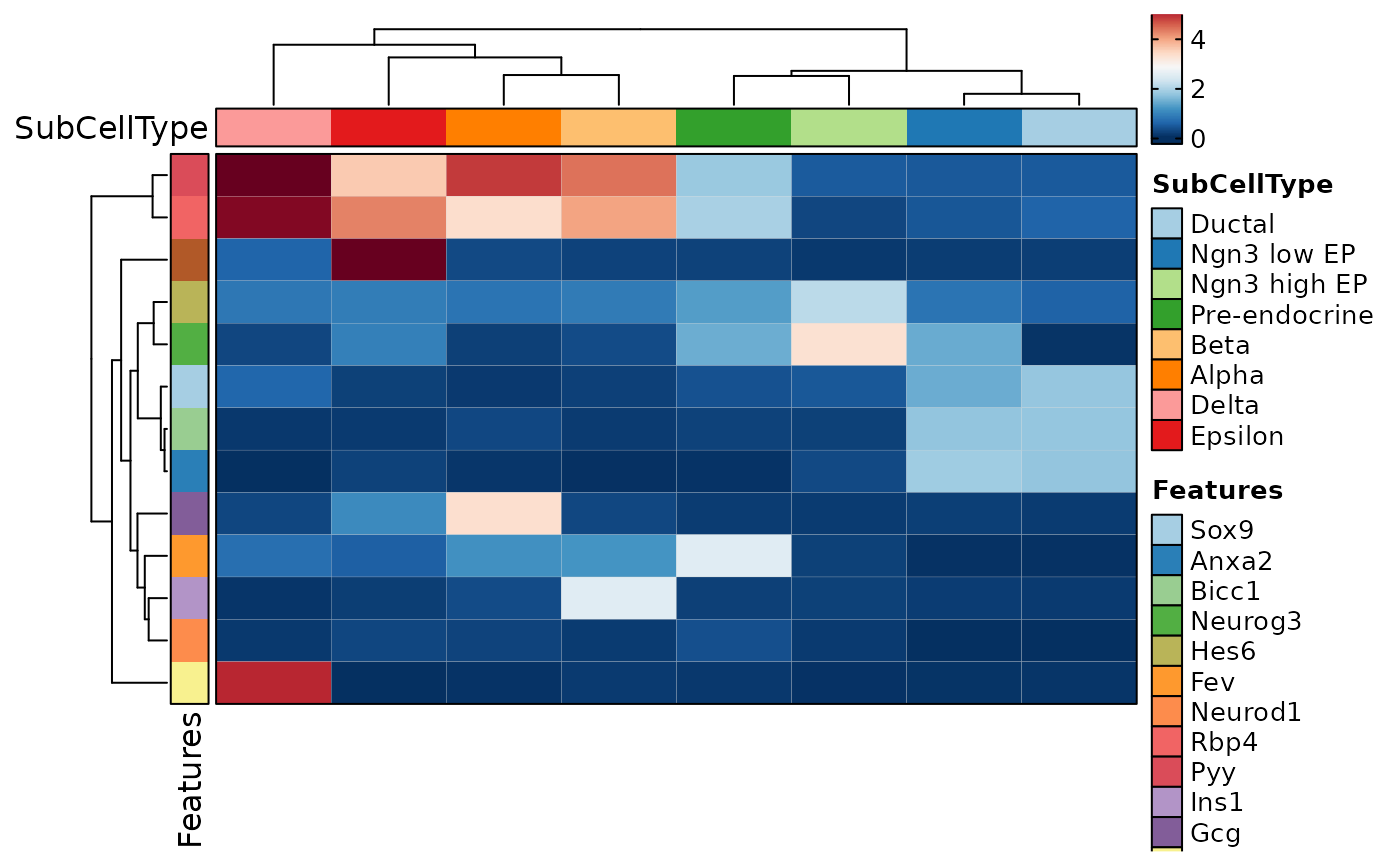 FeatureStatPlot(pancreas_sub, features = c("G2M_score", "nCount_RNA"),
ident = "SubCellType", plot_type = "dim", facet_by = "Phase", split_by = TRUE, ncol = 1)
FeatureStatPlot(pancreas_sub, features = c("G2M_score", "nCount_RNA"),
ident = "SubCellType", plot_type = "dim", facet_by = "Phase", split_by = TRUE, ncol = 1)
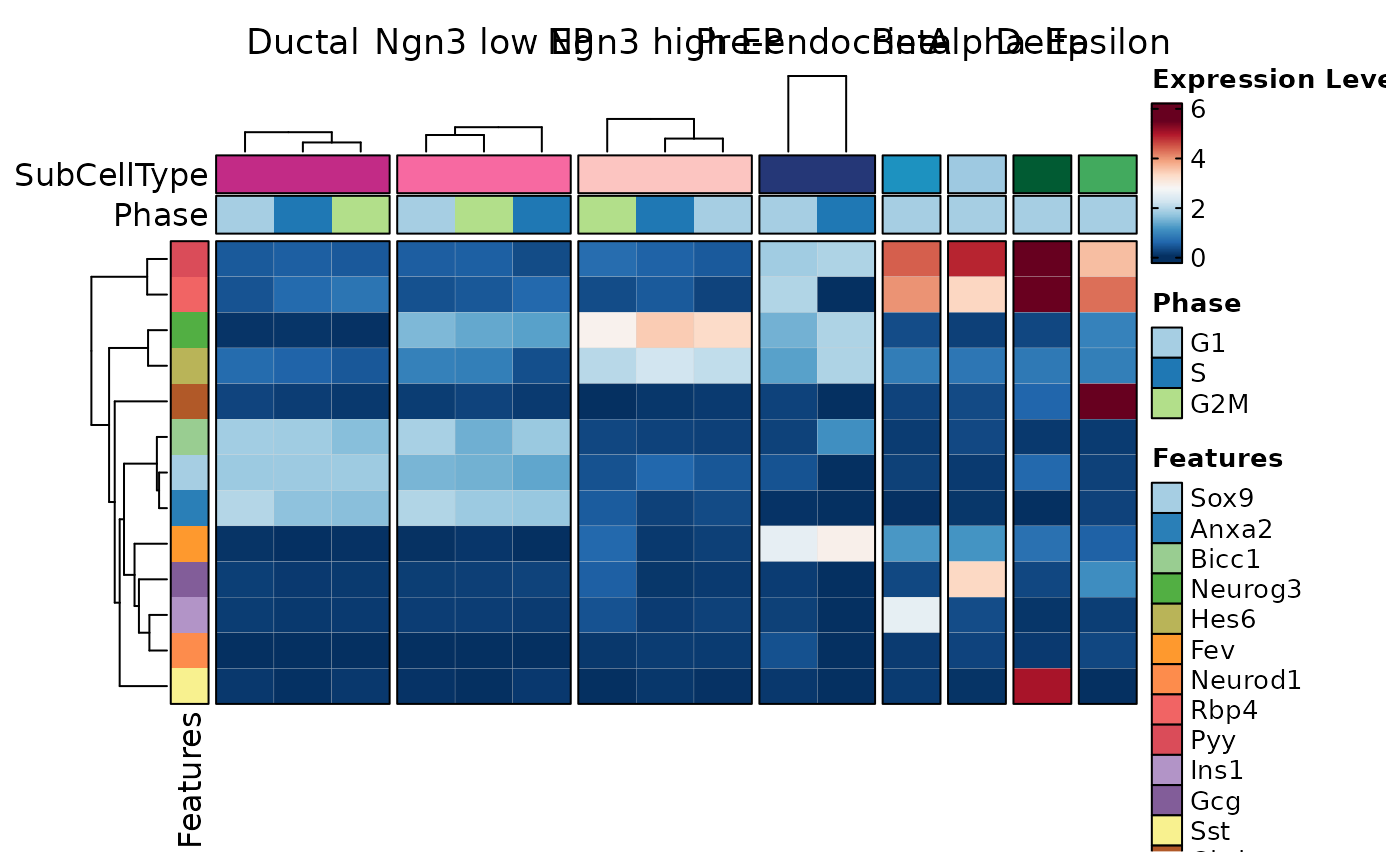 # Heatmap
features <- c(
"Sox9", "Anxa2", "Bicc1", # Ductal
"Neurog3", "Hes6", # EPs
"Fev", "Neurod1", # Pre-endocrine
"Rbp4", "Pyy", # Endocrine
"Ins1", "Gcg", "Sst", "Ghrl" # Beta, Alpha, Delta, Epsilon
)
rows_data <- data.frame(
Features = features, # 'rows_name' default is "Features"
group = c(
"Ductal", "Ductal", "Ductal", "EPs", "EPs", "Pre-endocrine",
"Pre-endocrine", "Endocrine", "Endocrine", "Beta", "Alpha", "Delta", "Epsilon"),
TF = c(TRUE, FALSE, FALSE, TRUE, FALSE, TRUE, FALSE, TRUE, FALSE, TRUE,
TRUE, TRUE, TRUE),
CSPA = c(FALSE, FALSE, FALSE, FALSE, TRUE, FALSE, FALSE, FALSE, TRUE, TRUE,
FALSE, FALSE, FALSE)
)
FeatureStatPlot(pancreas_sub, features = features, ident = "SubCellType",
plot_type = "heatmap", name = "Expression Level")
# Heatmap
features <- c(
"Sox9", "Anxa2", "Bicc1", # Ductal
"Neurog3", "Hes6", # EPs
"Fev", "Neurod1", # Pre-endocrine
"Rbp4", "Pyy", # Endocrine
"Ins1", "Gcg", "Sst", "Ghrl" # Beta, Alpha, Delta, Epsilon
)
rows_data <- data.frame(
Features = features, # 'rows_name' default is "Features"
group = c(
"Ductal", "Ductal", "Ductal", "EPs", "EPs", "Pre-endocrine",
"Pre-endocrine", "Endocrine", "Endocrine", "Beta", "Alpha", "Delta", "Epsilon"),
TF = c(TRUE, FALSE, FALSE, TRUE, FALSE, TRUE, FALSE, TRUE, FALSE, TRUE,
TRUE, TRUE, TRUE),
CSPA = c(FALSE, FALSE, FALSE, FALSE, TRUE, FALSE, FALSE, FALSE, TRUE, TRUE,
FALSE, FALSE, FALSE)
)
FeatureStatPlot(pancreas_sub, features = features, ident = "SubCellType",
plot_type = "heatmap", name = "Expression Level")
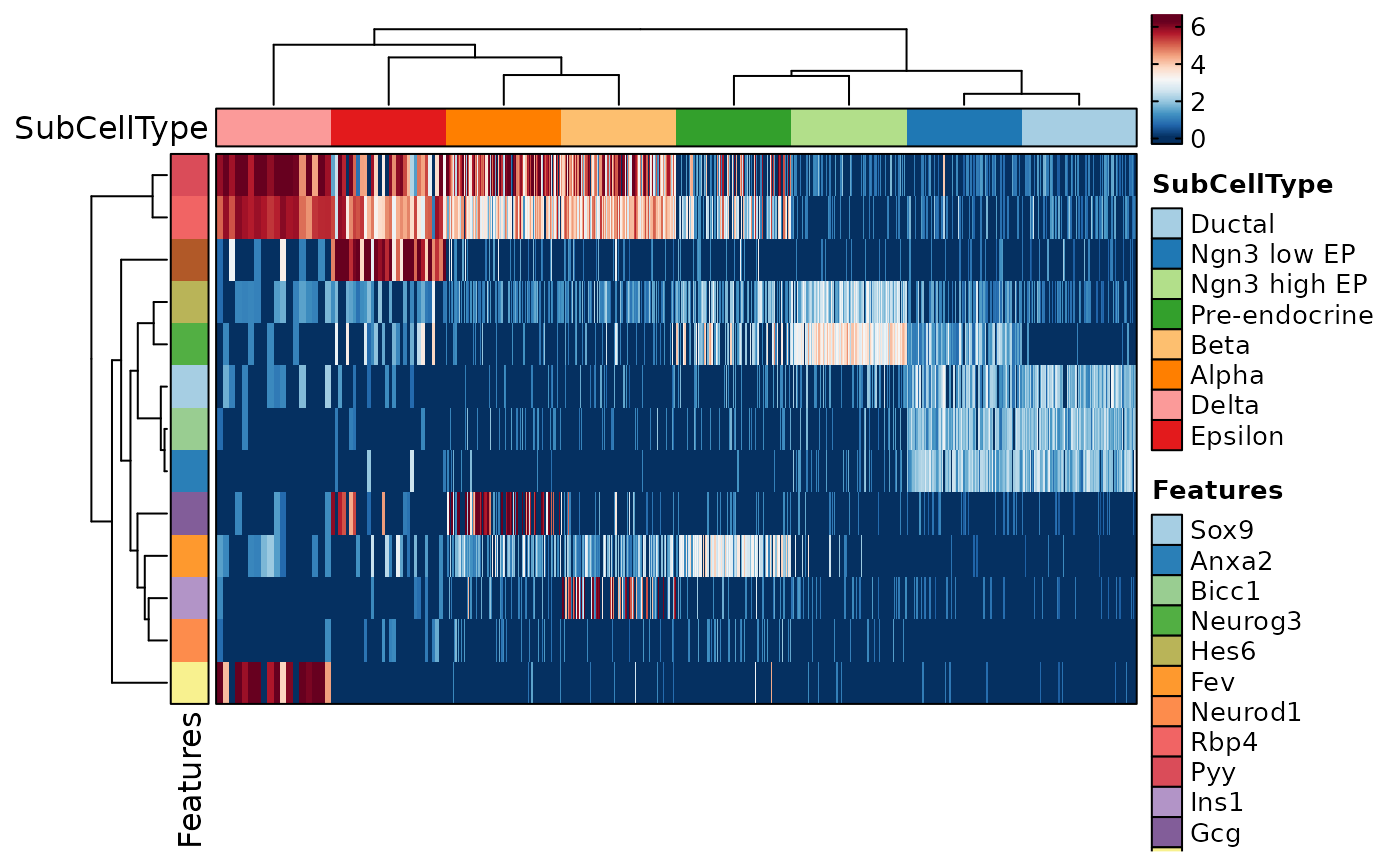 FeatureStatPlot(pancreas_sub, features = features, ident = "Phase",
plot_type = "heatmap", name = "Expression Level", columns_split_by = "SubCellType")
FeatureStatPlot(pancreas_sub, features = features, ident = "Phase",
plot_type = "heatmap", name = "Expression Level", columns_split_by = "SubCellType")
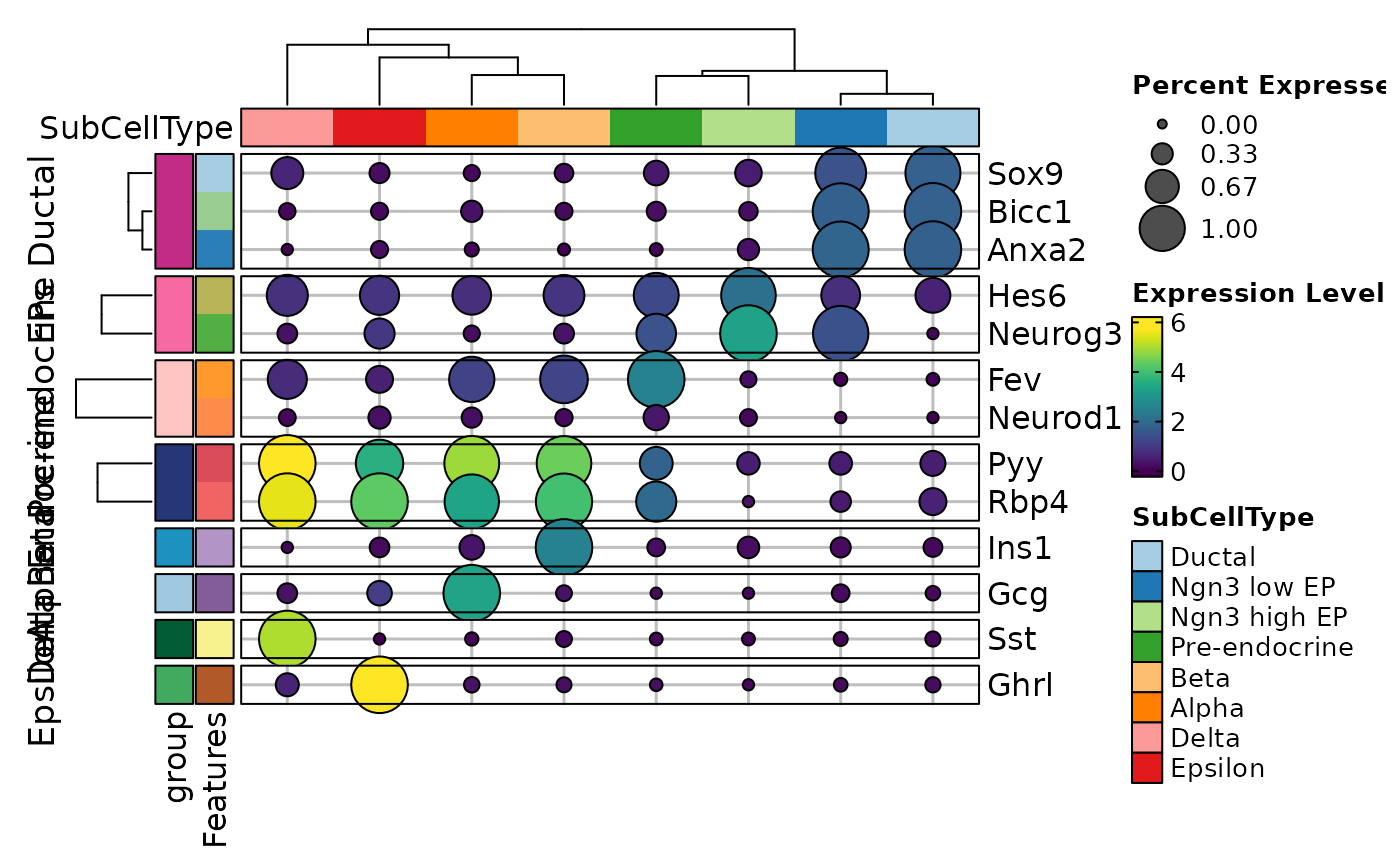
 FeatureStatPlot(pancreas_sub, features = features, ident = "SubCellType",
plot_type = "heatmap", cell_type = "bars", name = "Expression Level")
FeatureStatPlot(pancreas_sub, features = features, ident = "SubCellType",
plot_type = "heatmap", cell_type = "bars", name = "Expression Level")
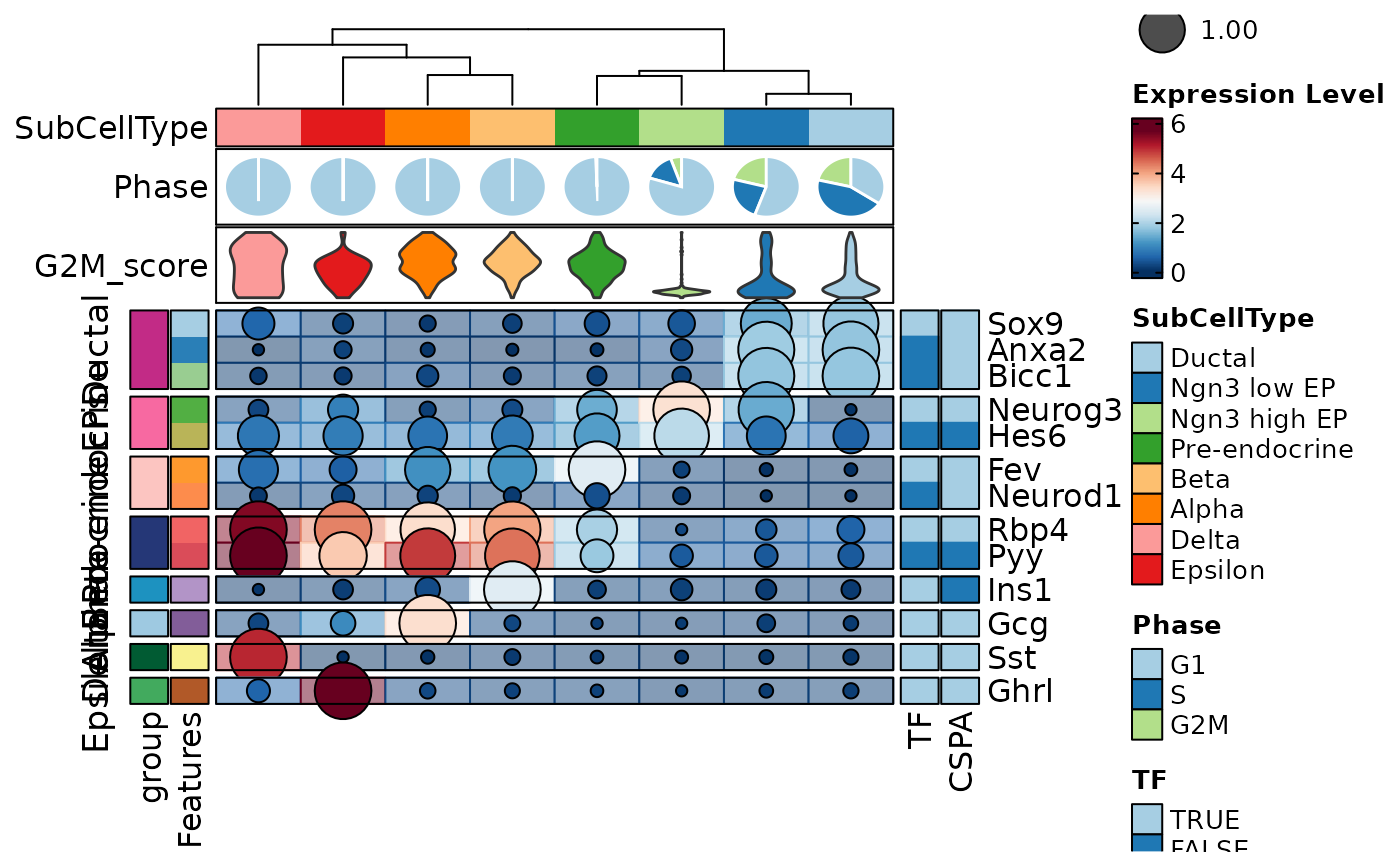
 FeatureStatPlot(pancreas_sub, features = features, ident = "SubCellType", cell_type = "dot",
plot_type = "heatmap", name = "Expression Level", dot_size = function(x) sum(x > 0) / length(x),
dot_size_name = "Percent Expressed", add_bg = TRUE, rows_data = rows_data,
show_row_names = TRUE, rows_split_by = "group", cluster_rows = FALSE,
column_annotation = c("Phase", "G2M_score"),
column_annotation_type = list(Phase = "pie", G2M_score = "violin"),
column_annotation_params = list(G2M_score = list(show_legend = FALSE)),
row_annotation = c("TF", "CSPA"),
row_annotation_side = "right",
row_annotation_type = list(TF = "simple", CSPA = "simple"))
#> Warning: [Heatmap] Assuming 'row_annotation_agg["TF"] = dplyr::first' for the simple annotation
#> Warning: [Heatmap] Assuming 'row_annotation_agg["CSPA"] = dplyr::first' for the simple annotation
FeatureStatPlot(pancreas_sub, features = features, ident = "SubCellType", cell_type = "dot",
plot_type = "heatmap", name = "Expression Level", dot_size = function(x) sum(x > 0) / length(x),
dot_size_name = "Percent Expressed", add_bg = TRUE, rows_data = rows_data,
show_row_names = TRUE, rows_split_by = "group", cluster_rows = FALSE,
column_annotation = c("Phase", "G2M_score"),
column_annotation_type = list(Phase = "pie", G2M_score = "violin"),
column_annotation_params = list(G2M_score = list(show_legend = FALSE)),
row_annotation = c("TF", "CSPA"),
row_annotation_side = "right",
row_annotation_type = list(TF = "simple", CSPA = "simple"))
#> Warning: [Heatmap] Assuming 'row_annotation_agg["TF"] = dplyr::first' for the simple annotation
#> Warning: [Heatmap] Assuming 'row_annotation_agg["CSPA"] = dplyr::first' for the simple annotation
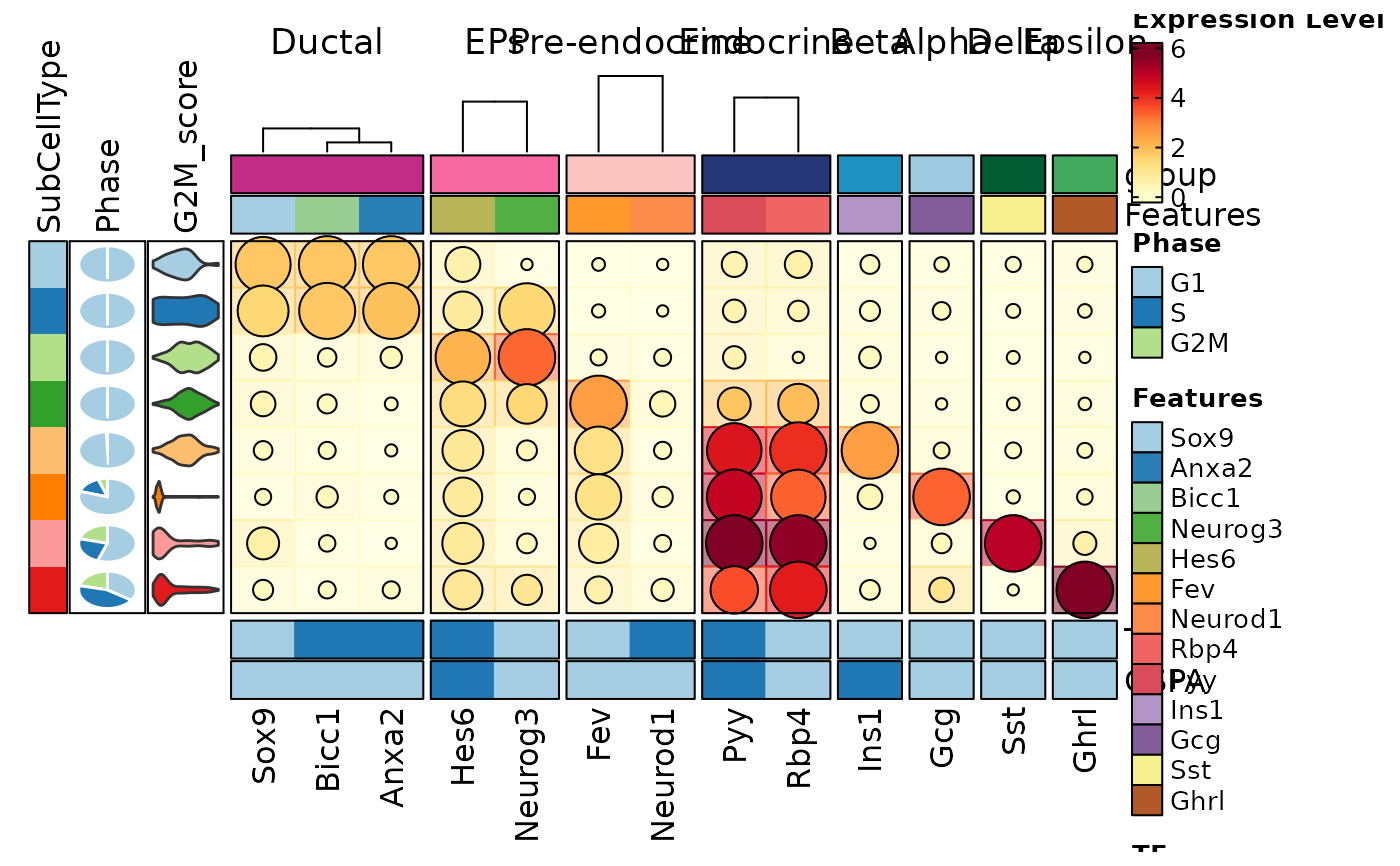
 FeatureStatPlot(pancreas_sub, features = features, ident = "SubCellType", cell_type = "dot",
plot_type = "heatmap", name = "Expression Level", dot_size = function(x) sum(x > 0) / length(x),
dot_size_name = "Percent Expressed", add_bg = TRUE,
rows_data = rows_data, show_column_names = TRUE, rows_split_by = "group",
cluster_rows = FALSE, flip = TRUE, palette = "YlOrRd",
column_annotation = c("Phase", "G2M_score"),
column_annotation_type = list(Phase = "pie", G2M_score = "violin"),
column_annotation_params = list(G2M_score = list(show_legend = FALSE)),
row_annotation = c("TF", "CSPA"),
row_annotation_side = "right",
row_annotation_type = list(TF = "simple", CSPA = "simple"))
#> Warning: [Heatmap] Assuming 'row_annotation_agg["TF"] = dplyr::first' for the simple annotation
#> Warning: [Heatmap] Assuming 'row_annotation_agg["CSPA"] = dplyr::first' for the simple annotation
FeatureStatPlot(pancreas_sub, features = features, ident = "SubCellType", cell_type = "dot",
plot_type = "heatmap", name = "Expression Level", dot_size = function(x) sum(x > 0) / length(x),
dot_size_name = "Percent Expressed", add_bg = TRUE,
rows_data = rows_data, show_column_names = TRUE, rows_split_by = "group",
cluster_rows = FALSE, flip = TRUE, palette = "YlOrRd",
column_annotation = c("Phase", "G2M_score"),
column_annotation_type = list(Phase = "pie", G2M_score = "violin"),
column_annotation_params = list(G2M_score = list(show_legend = FALSE)),
row_annotation = c("TF", "CSPA"),
row_annotation_side = "right",
row_annotation_type = list(TF = "simple", CSPA = "simple"))
#> Warning: [Heatmap] Assuming 'row_annotation_agg["TF"] = dplyr::first' for the simple annotation
#> Warning: [Heatmap] Assuming 'row_annotation_agg["CSPA"] = dplyr::first' for the simple annotation
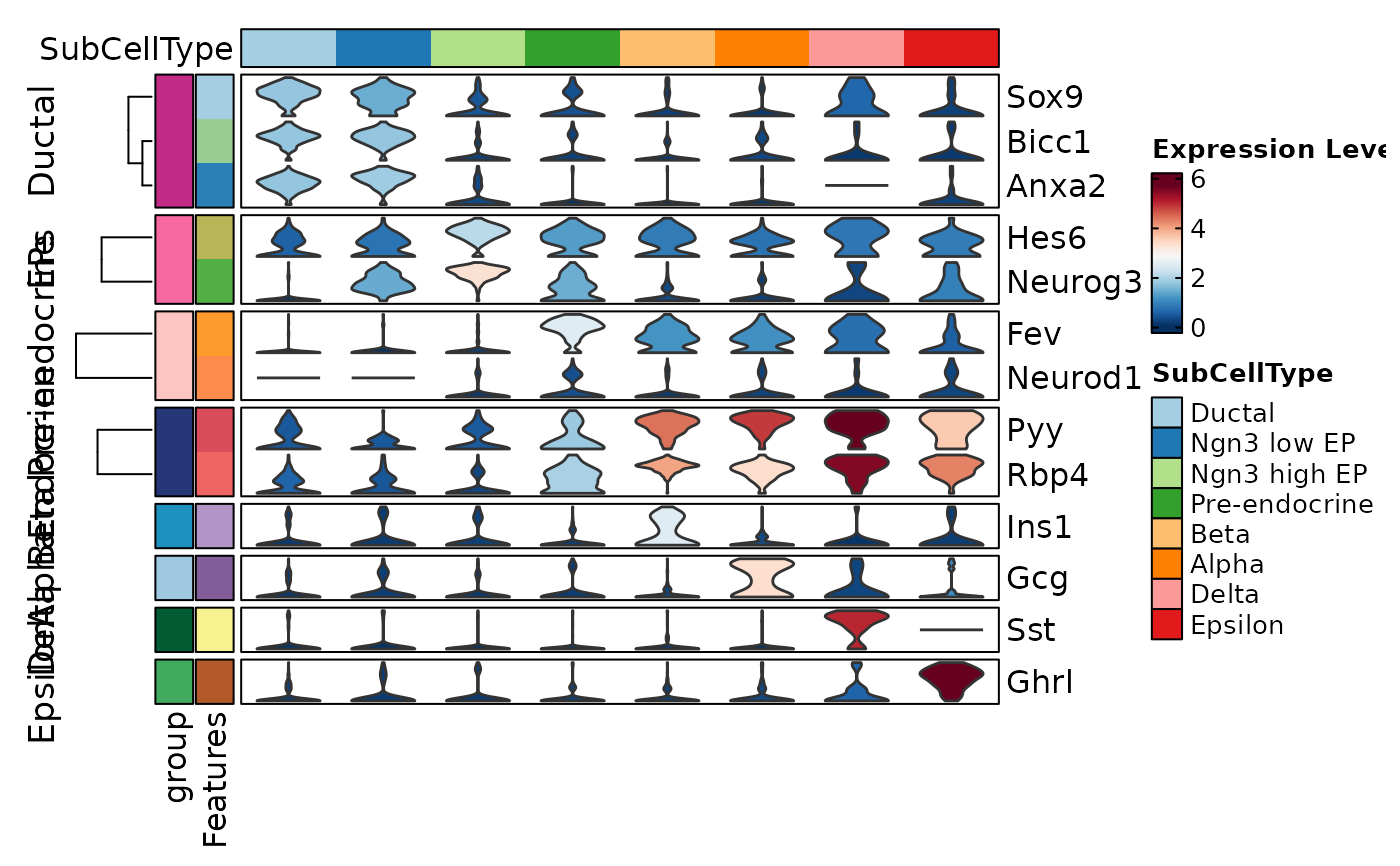
 FeatureStatPlot(pancreas_sub, features = features, ident = "SubCellType", cell_type = "violin",
plot_type = "heatmap", name = "Expression Level", show_row_names = TRUE,
cluster_columns = FALSE, rows_split_by = "group", rows_data = rows_data)
FeatureStatPlot(pancreas_sub, features = features, ident = "SubCellType", cell_type = "violin",
plot_type = "heatmap", name = "Expression Level", show_row_names = TRUE,
cluster_columns = FALSE, rows_split_by = "group", rows_data = rows_data)
 FeatureStatPlot(pancreas_sub, features = features, ident = "SubCellType", cell_type = "dot",
plot_type = "heatmap", dot_size = function(x) sum(x > 0) / length(x),
dot_size_name = "Percent Expressed", palette = "viridis", add_reticle = TRUE,
rows_data = rows_data, name = "Expression Level", show_row_names = TRUE,
rows_split_by = "group")
FeatureStatPlot(pancreas_sub, features = features, ident = "SubCellType", cell_type = "dot",
plot_type = "heatmap", dot_size = function(x) sum(x > 0) / length(x),
dot_size_name = "Percent Expressed", palette = "viridis", add_reticle = TRUE,
rows_data = rows_data, name = "Expression Level", show_row_names = TRUE,
rows_split_by = "group")
 # Visualize the markers for each sub-cell type (the markers can overlap)
# Say: markers <- Seurat::FindAllMarkers(pancreas_sub, ident = "SubCellType")
markers <- data.frame(
avg_log2FC = c(
3.44, 2.93, 2.72, 2.63, 2.13, 1.97, 2.96, 1.92, 5.22, 3.91, 3.64, 4.52,
3.45, 2.45, 1.75, 2.08, 9.10, 4.45, 3.61, 6.30, 4.96, 3.49, 3.91, 3.90,
10.58, 5.84, 4.73, 3.34, 7.22, 4.52, 10.10, 4.25),
cluster = factor(rep(
c("Ductal", "Ngn3 low EP", "Ngn3 high EP", "Pre-endocrine", "Beta",
"Alpha", "Delta", "Epsilon"), each = 4),
levels = levels(pancreas_sub$SubCellType)),
gene = c(
"Cyr61", "Adamts1", "Anxa2", "Bicc1", "1700011H14Rik", "Gsta3", "8430408G22Rik",
"Anxa2", "Ppp1r14a", "Btbd17", "Neurog3", "Gadd45a", "Fev", "Runx1t1", "Hmgn3",
"Cryba2", "Ins2", "Ppp1r1a", "Gng12", "Sytl4", "Irx1", "Tmem27", "Peg10", "Irx2",
"Sst", "Ptprz1", "Arg1", "Frzb", "Irs4", "Mboat4", "Ghrl", "Arg1"
)
)
FeatureStatPlot(pancreas_sub,
features = unique(markers$gene), ident = "SubCellType", cell_type = "bars",
plot_type = "heatmap", rows_data = markers, rows_name = "gene", rows_split_by = "cluster",
show_row_names = TRUE, show_column_names = TRUE, name = "Expression Level",
cluster_rows = FALSE, cluster_columns = FALSE, rows_split_palette = "Paired")
# Visualize the markers for each sub-cell type (the markers can overlap)
# Say: markers <- Seurat::FindAllMarkers(pancreas_sub, ident = "SubCellType")
markers <- data.frame(
avg_log2FC = c(
3.44, 2.93, 2.72, 2.63, 2.13, 1.97, 2.96, 1.92, 5.22, 3.91, 3.64, 4.52,
3.45, 2.45, 1.75, 2.08, 9.10, 4.45, 3.61, 6.30, 4.96, 3.49, 3.91, 3.90,
10.58, 5.84, 4.73, 3.34, 7.22, 4.52, 10.10, 4.25),
cluster = factor(rep(
c("Ductal", "Ngn3 low EP", "Ngn3 high EP", "Pre-endocrine", "Beta",
"Alpha", "Delta", "Epsilon"), each = 4),
levels = levels(pancreas_sub$SubCellType)),
gene = c(
"Cyr61", "Adamts1", "Anxa2", "Bicc1", "1700011H14Rik", "Gsta3", "8430408G22Rik",
"Anxa2", "Ppp1r14a", "Btbd17", "Neurog3", "Gadd45a", "Fev", "Runx1t1", "Hmgn3",
"Cryba2", "Ins2", "Ppp1r1a", "Gng12", "Sytl4", "Irx1", "Tmem27", "Peg10", "Irx2",
"Sst", "Ptprz1", "Arg1", "Frzb", "Irs4", "Mboat4", "Ghrl", "Arg1"
)
)
FeatureStatPlot(pancreas_sub,
features = unique(markers$gene), ident = "SubCellType", cell_type = "bars",
plot_type = "heatmap", rows_data = markers, rows_name = "gene", rows_split_by = "cluster",
show_row_names = TRUE, show_column_names = TRUE, name = "Expression Level",
cluster_rows = FALSE, cluster_columns = FALSE, rows_split_palette = "Paired")
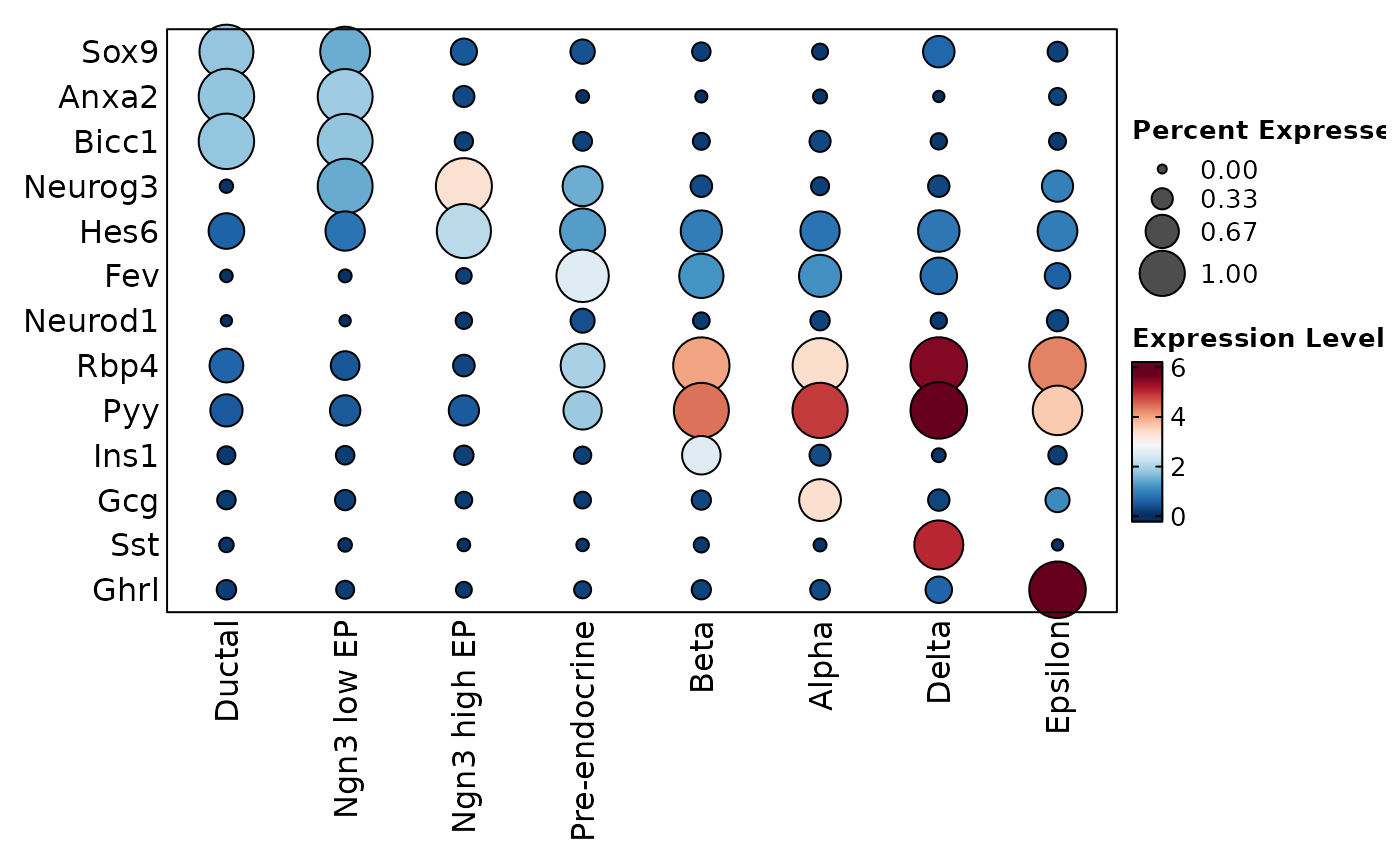
 # Use plot_type = "dot" to as a shortcut for heatmap with cell_type = "dot"
FeatureStatPlot(pancreas_sub, features = features, ident = "SubCellType", plot_type = "dot")
# Use plot_type = "dot" to as a shortcut for heatmap with cell_type = "dot"
FeatureStatPlot(pancreas_sub, features = features, ident = "SubCellType", plot_type = "dot")
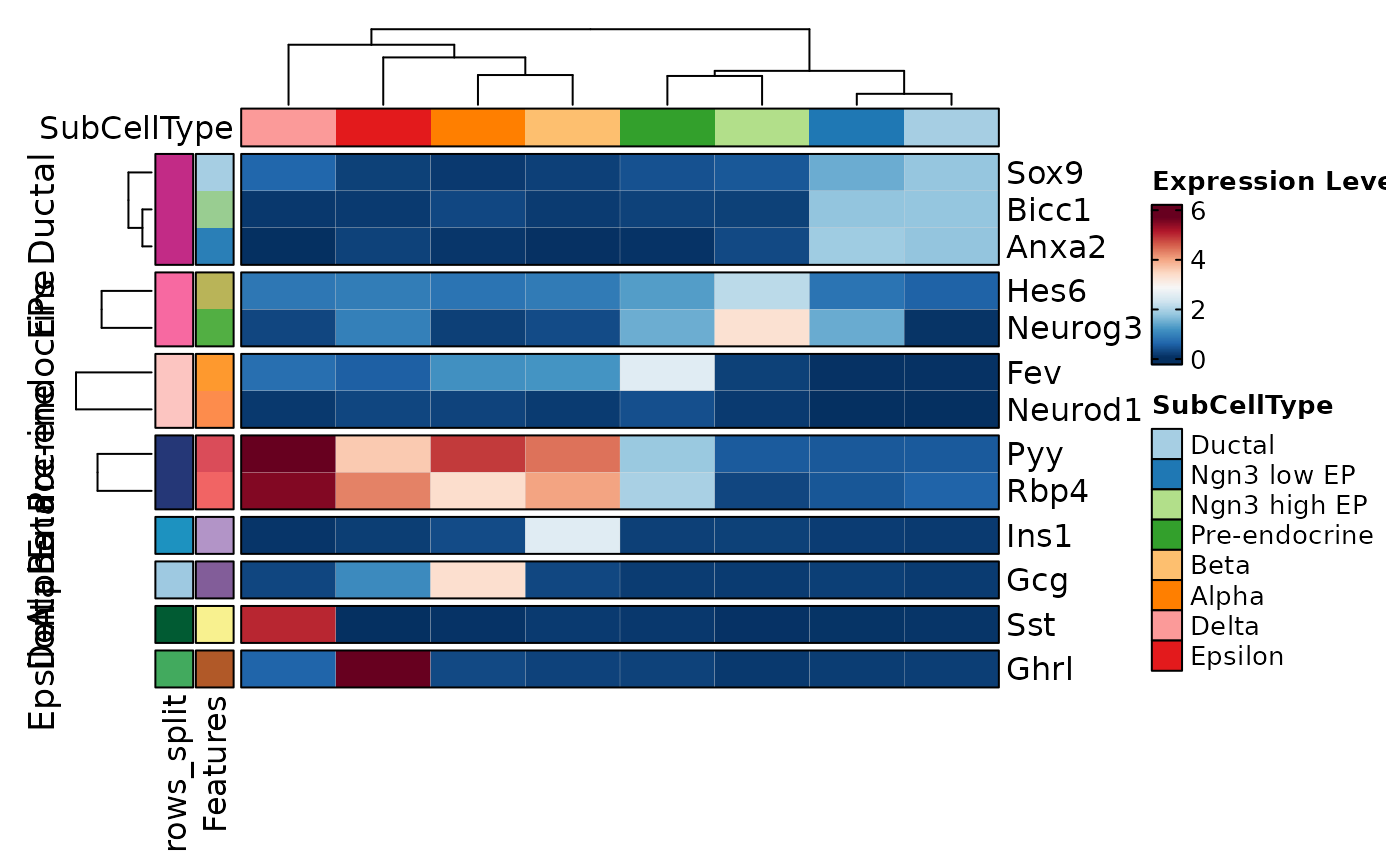
 named_features <- list(
Ductal = c("Sox9", "Anxa2", "Bicc1"),
EPs = c("Neurog3", "Hes6"),
`Pre-endocrine` = c("Fev", "Neurod1"),
Endocrine = c("Rbp4", "Pyy"),
Beta = "Ins1", Alpha = "Gcg", Delta = "Sst", Epsilon = "Ghrl"
)
FeatureStatPlot(pancreas_sub, features = named_features, ident = "SubCellType",
plot_type = "heatmap", name = "Expression Level", show_row_names = TRUE)
named_features <- list(
Ductal = c("Sox9", "Anxa2", "Bicc1"),
EPs = c("Neurog3", "Hes6"),
`Pre-endocrine` = c("Fev", "Neurod1"),
Endocrine = c("Rbp4", "Pyy"),
Beta = "Ins1", Alpha = "Gcg", Delta = "Sst", Epsilon = "Ghrl"
)
FeatureStatPlot(pancreas_sub, features = named_features, ident = "SubCellType",
plot_type = "heatmap", name = "Expression Level", show_row_names = TRUE)
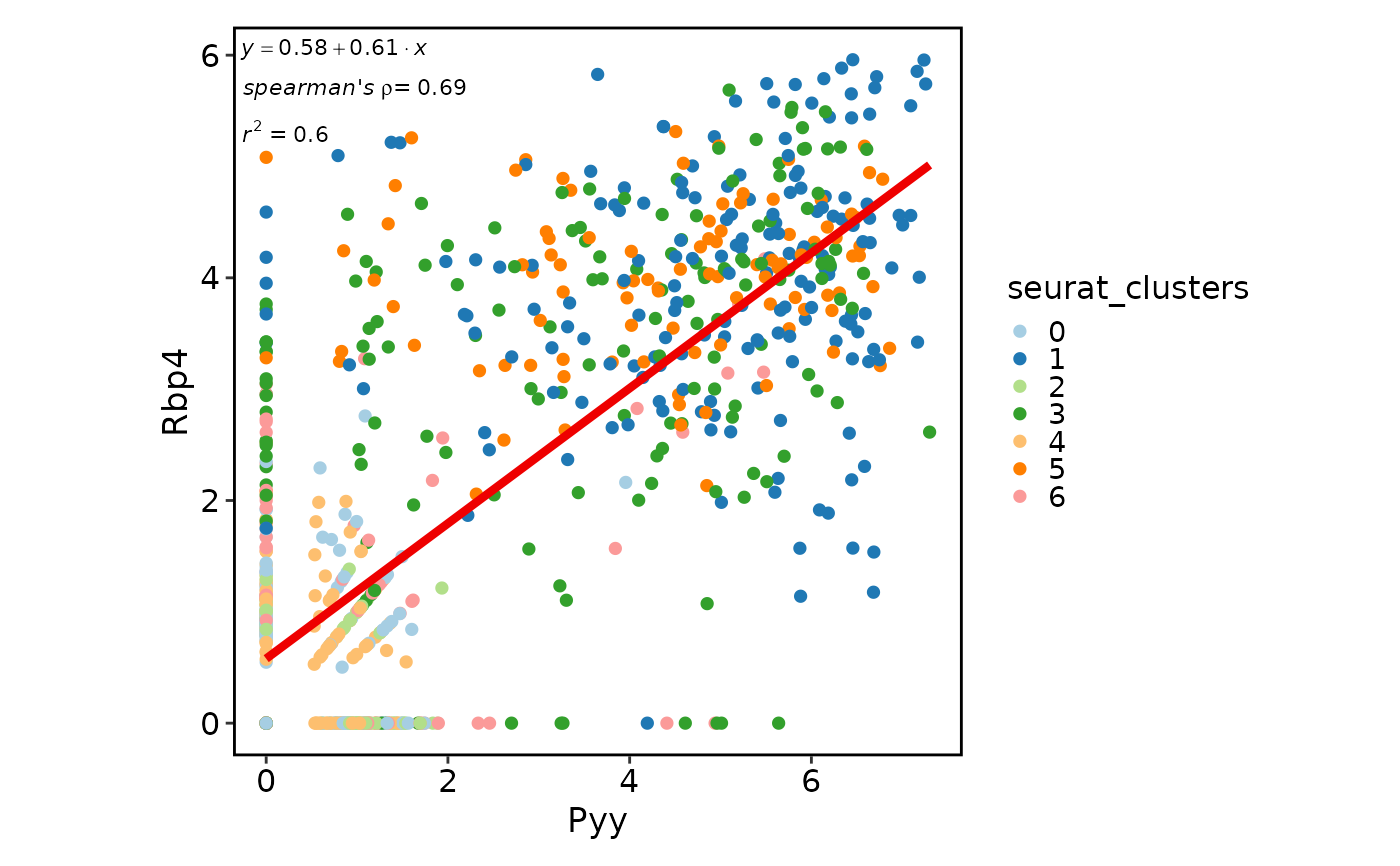
 # Correlation plot
FeatureStatPlot(pancreas_sub, features = c("Pyy", "Rbp4"), plot_type = "cor",
anno_items = c("eq", "r2", "spearman"))
# Correlation plot
FeatureStatPlot(pancreas_sub, features = c("Pyy", "Rbp4"), plot_type = "cor",
anno_items = c("eq", "r2", "spearman"))
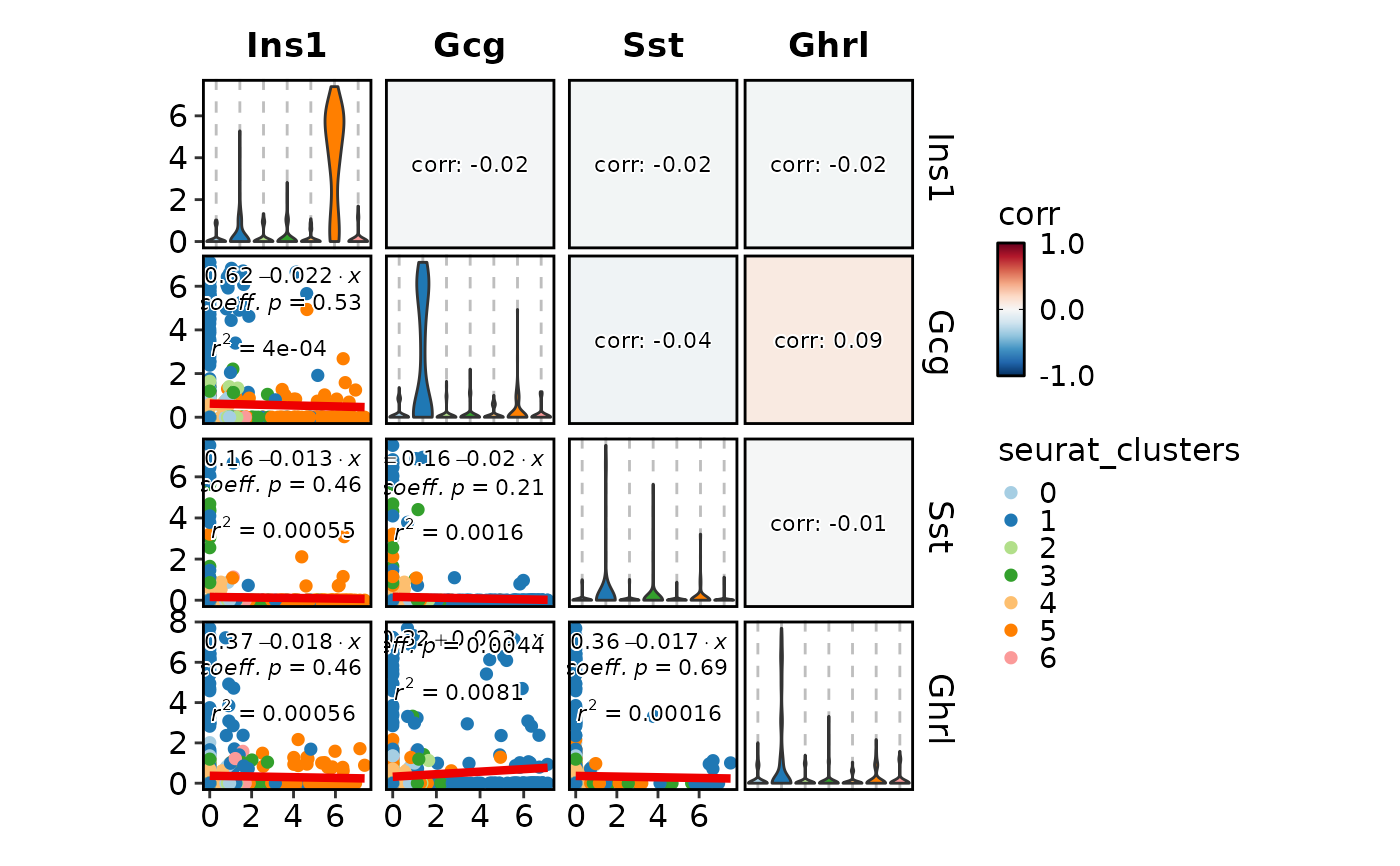
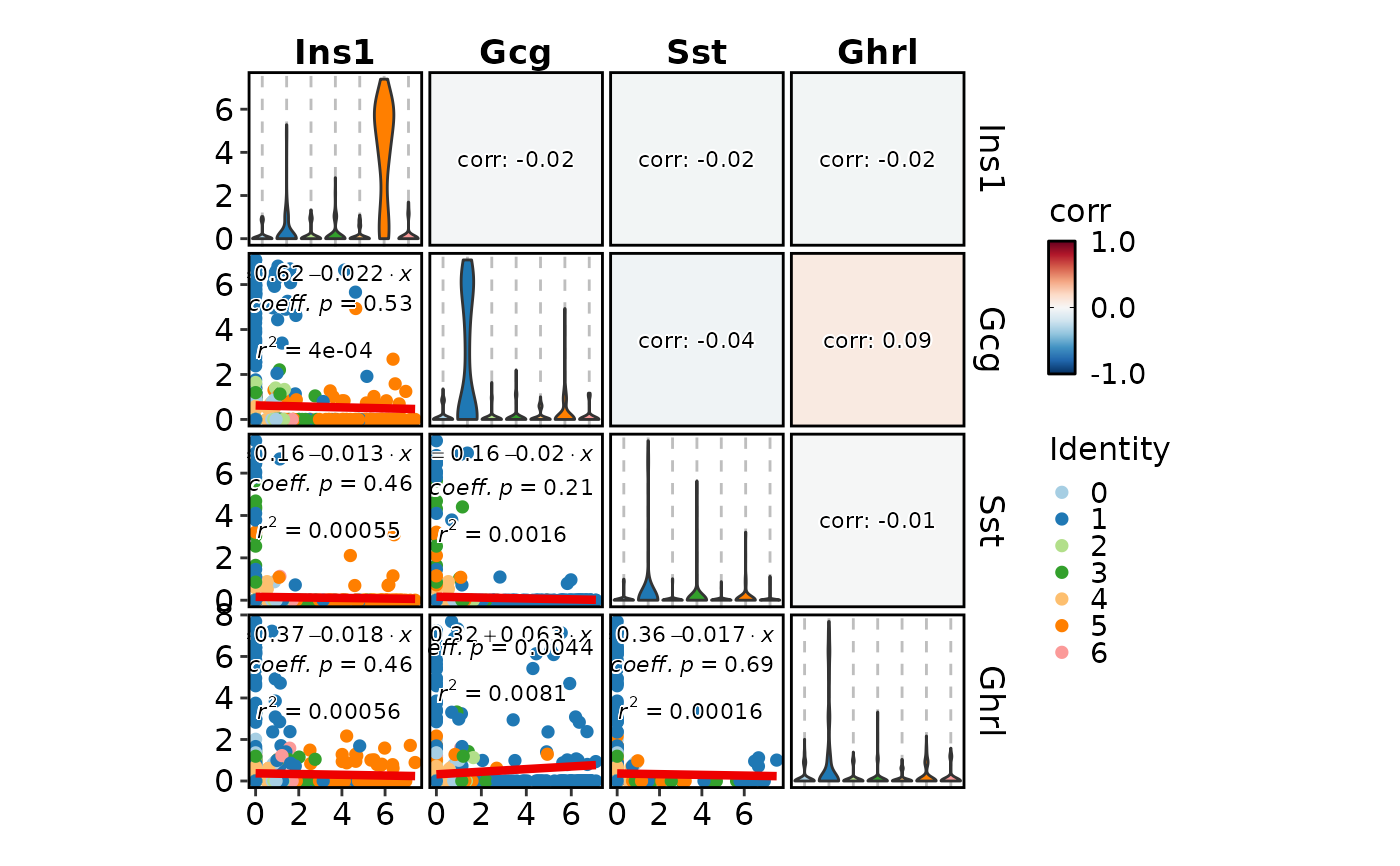
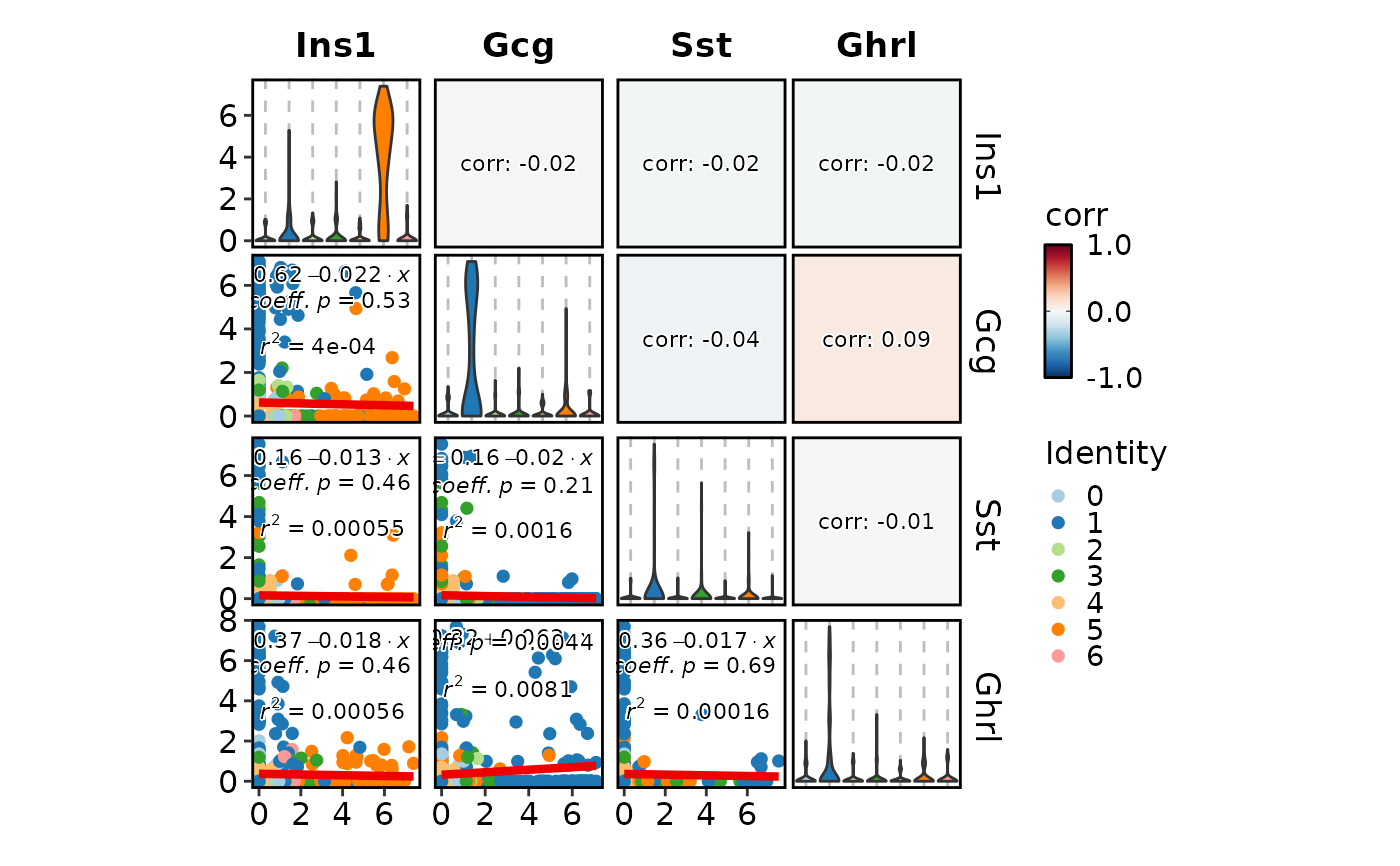 FeatureStatPlot(pancreas_sub, features = c("Ins1", "Gcg", "Sst", "Ghrl"),
plot_type = "cor")
FeatureStatPlot(pancreas_sub, features = c("Ins1", "Gcg", "Sst", "Ghrl"),
plot_type = "cor")
 # }
# }