Plot markers, typically identified by Seurat::FindMarkers() or Seurat::FindAllMarkers().
Usage
MarkersPlot(
markers,
object = NULL,
plot_type = c("volcano", "volcano_log2fc", "volcano_pct", "jitter", "jitter_log2fc",
"jitter_pct", "heatmap_log2fc", "heatmap_pct", "dot_log2fc", "dot_pct", "heatmap",
"violin", "box", "bar", "ridge", "dot"),
subset_by = NULL,
subset_as_facet = FALSE,
comparison_by = NULL,
p_adjust = TRUE,
cutoff = NULL,
order_by = NULL,
select = ifelse(plot_type %in% c("volcano", "volcano_log2fc", "volcano_pct",
"jitter", "jitter_log2fc", "jitter_pct"), 5, 10),
...
)Arguments
- markers
A data frame of markers, typically identified by
Seurat::FindMarkers()orSeurat::FindAllMarkers().- object
A Seurat object. Required for some plot types, see
plot_type.- plot_type
Type of plot to generate. Options include:
volcano/volcano_log2fc: Volcano plot with log2 fold change on x-axis and -log10(p-value) on y-axis. Ifp_adjustis TRUE, -log10(adjusted p-value) is used instead.volcano_pct: Volcano plot with difference in percentage of cells expressing the gene between two groups on x-axis and -log10(p-value) on y-axis. Ifp_adjustis TRUE, -log10(adjusted p-value) is used instead.jitter/jitter_log2fc: Jitter plot of log2 fold change for each gene. The x-axis is the groups defined bysubset_by, and the y-axis is log2 fold change. The size of the dots represents -log10(p-value) or -log10(adjusted p-value) ifp_adjustis TRUE.jitter_pct: Jitter plot of difference in percentage of cells expressing the gene between two groups for each gene. The x-axis is the groups defined bysubset_by, and the y-axis is the difference in percentage of cells expressing the gene between two groups. The size of the dots represents -log10(p-value) or -log10(adjusted p-value) ifp_adjustis TRUE.heatmap_log2fc: Heatmap of log2 fold change for each gene across groups defined bysubset_by. By specifyingcutoff, The heatmap cells will be labeled with "*" for p-value < cutoff, ifp_adjustis TRUE, adjusted p-value < cutoff.heatmap_pct: Heatmap of difference in percentage of cells expressing the gene between two groups for each gene across groups defined bysubset_by. By specifyingcutoff, The heatmap cells will be labeled with "*" for p-value < cutoff, ifp_adjustis TRUE, adjusted p-value < cutoff.dot_log2fc: Dot plot of log2 fold change for each gene across groups defined bysubset_by. The size of the dots represents -log10(p-value) or -log10(adjusted p-value) ifp_adjustis TRUE.dot_pct: Dot plot of difference in percentage of cells expressing the gene between two groups for each gene across groups defined bysubset_by. The size of the dots represents -log10(p-value) or -log10(adjusted p-value) ifp_adjustis TRUE.heatmap: Heatmap of expression values for each gene across groups defined bysubset_by. Requiresobject.violin: Violin plot of expression values for each gene across groups defined bysubset_by. Requiresobject.box: Box plot of expression values for each gene across groups defined bysubset_by. Requiresobject.bar: Bar plot of average expression values for each gene across groups defined bysubset_by. Requiresobject.ridge: Ridge plot of expression values for each gene across groups defined bysubset_by. Requiresobject.dot: Dot plot of expression values for each gene across groups defined bysubset_by. Requiresobject.
- subset_by
A column in markers indicating where the markers are identified from, e.g., cluster or condition. If object is provided, you can provide the corresponding metadata column in
subset_byto merge the markers with the object's metadata, using:as separator. For example, if the markers are identified from different clusters (identified byFindAllMarkers), and the object's ident column is "RNA_snn_res.0.8", you can setsubset_by = "cluster:RNA_snn_res.0.8". Note that for other columns to be merged, only the first value of each group defined bysubset_bywill be used. For some plots, this is used to split the markers into multiple plots, e.g., one plot for each cluster. Seeplot_typefor details.- subset_as_facet
Logical, whether to facet the plots by
subset_byif applicable.- comparison_by
The metadata column in markers indicating the comparion. When visualizing the expression values, this column should also be in the object's metadata. The values of this column should either a single value, indicating the comparison is between this group and all other cells (in the subset), Similar as
subset_by, you can provide the corresponding object metadata column by using:as separator. IfNULL, all markers are treated as from one comparison.- p_adjust
Logical, whether to use adjusted p-value for plots that involve p-values. Default is TRUE.
- cutoff
Numeric, p-value or adjusted p-value cutoff to label significance in heatmap plots. Default is NULL, no cutoff.
- order_by
A string of expression to order the markers within each group defined by
subset_by. In addition to the columns inmarkers, you can also use the columns from the object's metadata ifobjectis provided. The object's metadata will be merged withmarkersbysubset_by. Be carefull that only the first value of other columns will be used.- select
Number of top markers (ordered by
order_by) to select for each group defined bysubset_byor a string of expression to filter markers. It will be evaluated bydplyr::filter(). Forvolcano,volcano_log2fc,volcano_pct,jitter,jitter_log2fc, andjitter_pctplots, the selected markers will be labeled in the plot. FOr other plot types, only the selected markers will be plotted. Default is 5 forvolcano,volcano_log2fc,volcano_pct,jitter,jitter_log2fc,jitter_pct, and 10 for other plot types.- ...
Additional arguments passed to specific plotting functions. See Details.
Details
Additional arguments passed to specific plotting functions:
For
heatmap_log2fc,heatmap_pct,dot_log2fc, anddot_pct, the arguments will be passed toplotthis::Heatmap()For
violin, "box", "bar", "ridge" and "dot", the arguments will be passed toFeatureStatPlot()For
volcano,volcano_log2fc,volcano_pct, the arguments will be passed toplotthis::VolcanoPlot()For
jitter,jitter_log2fc,jitter_pct, the arguments will be passed toplotthis::JitterPlot()
Examples
# \donttest{
data(pancreas_sub)
markers <- Seurat::FindMarkers(pancreas_sub,
group.by = "Phase", ident.1 = "G2M", ident.2 = "G1")
#> For a (much!) faster implementation of the Wilcoxon Rank Sum Test,
#> (default method for FindMarkers) please install the presto package
#> --------------------------------------------
#> install.packages('devtools')
#> devtools::install_github('immunogenomics/presto')
#> --------------------------------------------
#> After installation of presto, Seurat will automatically use the more
#> efficient implementation (no further action necessary).
#> This message will be shown once per session
allmarkers <- Seurat::FindAllMarkers(pancreas_sub) # seurat_clusters
#> Calculating cluster 0
#> Calculating cluster 1
#> Calculating cluster 2
#> Calculating cluster 3
#> Calculating cluster 4
#> Calculating cluster 5
#> Calculating cluster 6
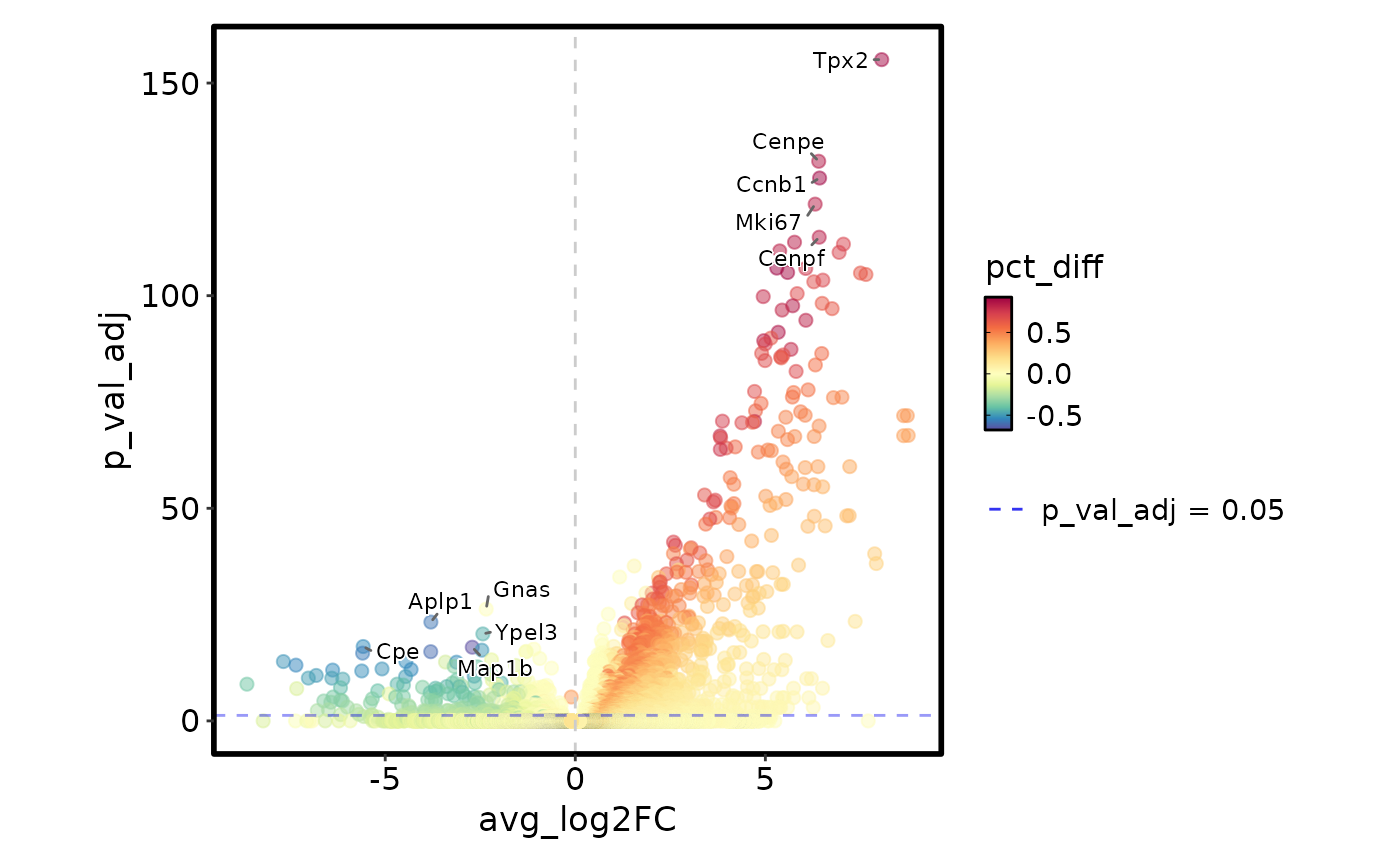
MarkersPlot(markers)
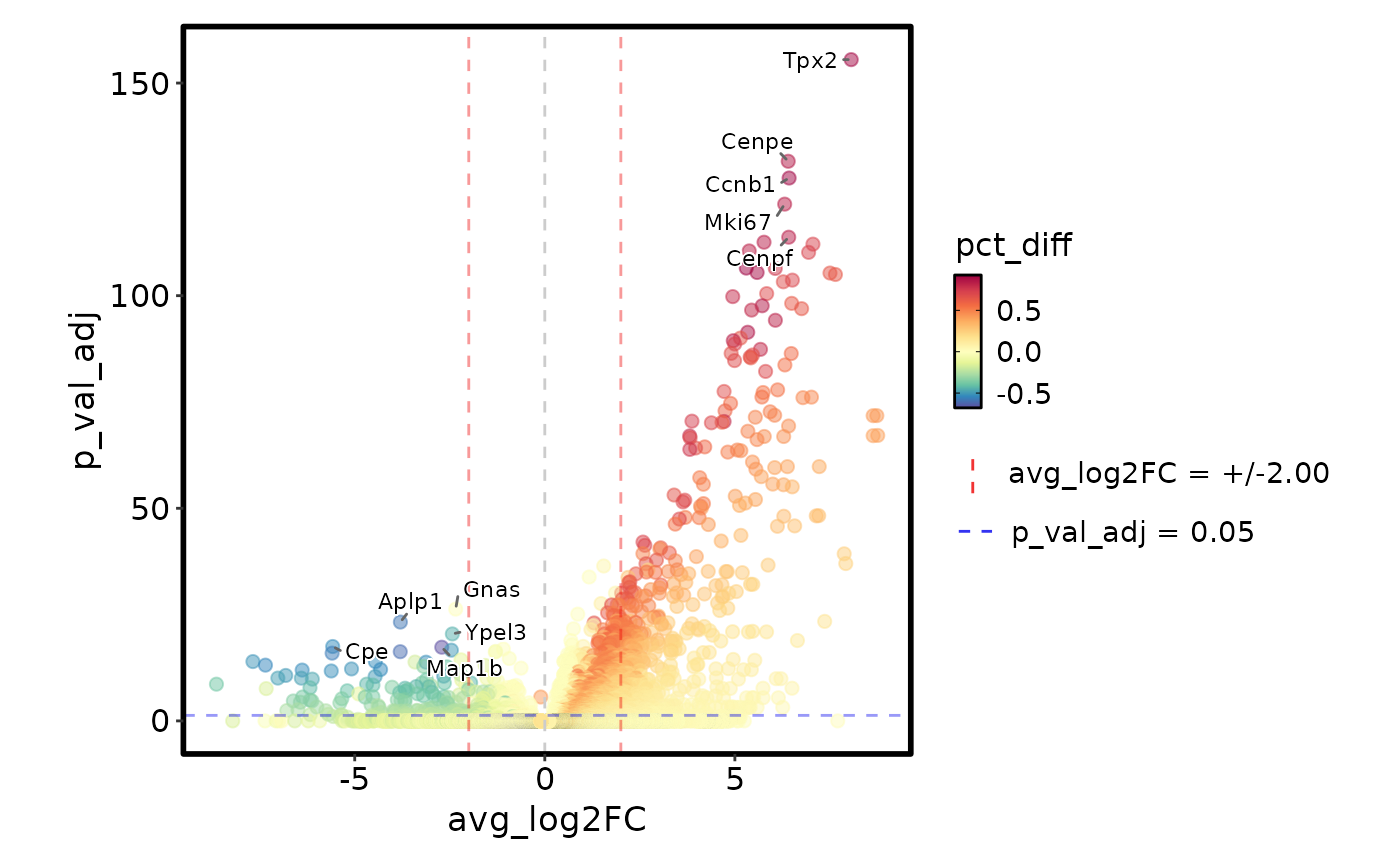
 MarkersPlot(markers, x_cutoff = 2)
MarkersPlot(markers, x_cutoff = 2)
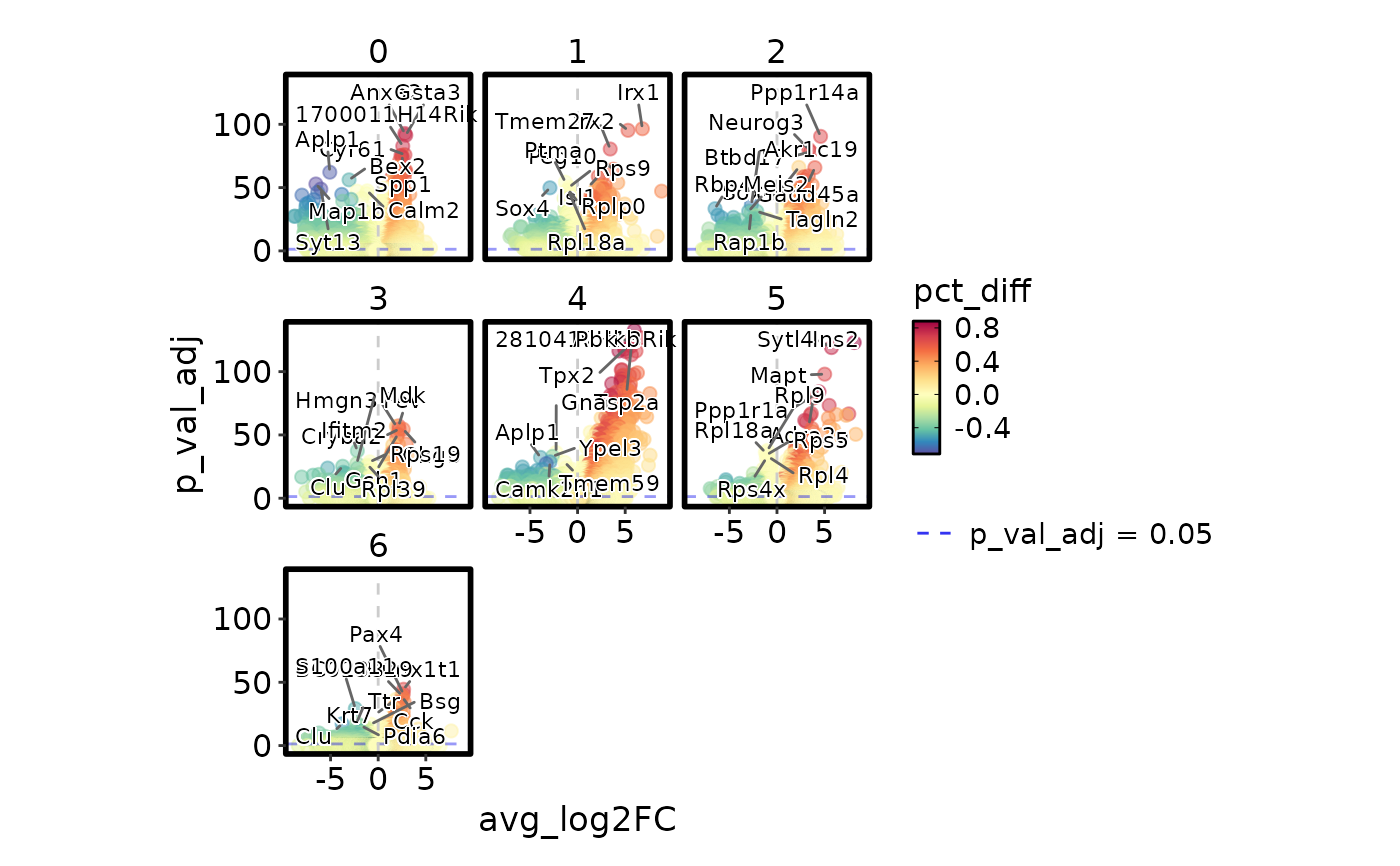
 MarkersPlot(allmarkers,
subset_by = "cluster", ncol = 2, subset_as_facet = TRUE)
MarkersPlot(allmarkers,
subset_by = "cluster", ncol = 2, subset_as_facet = TRUE)
 MarkersPlot(markers, plot_type = "volcano_pct", flip_negative = TRUE)
MarkersPlot(markers, plot_type = "volcano_pct", flip_negative = TRUE)
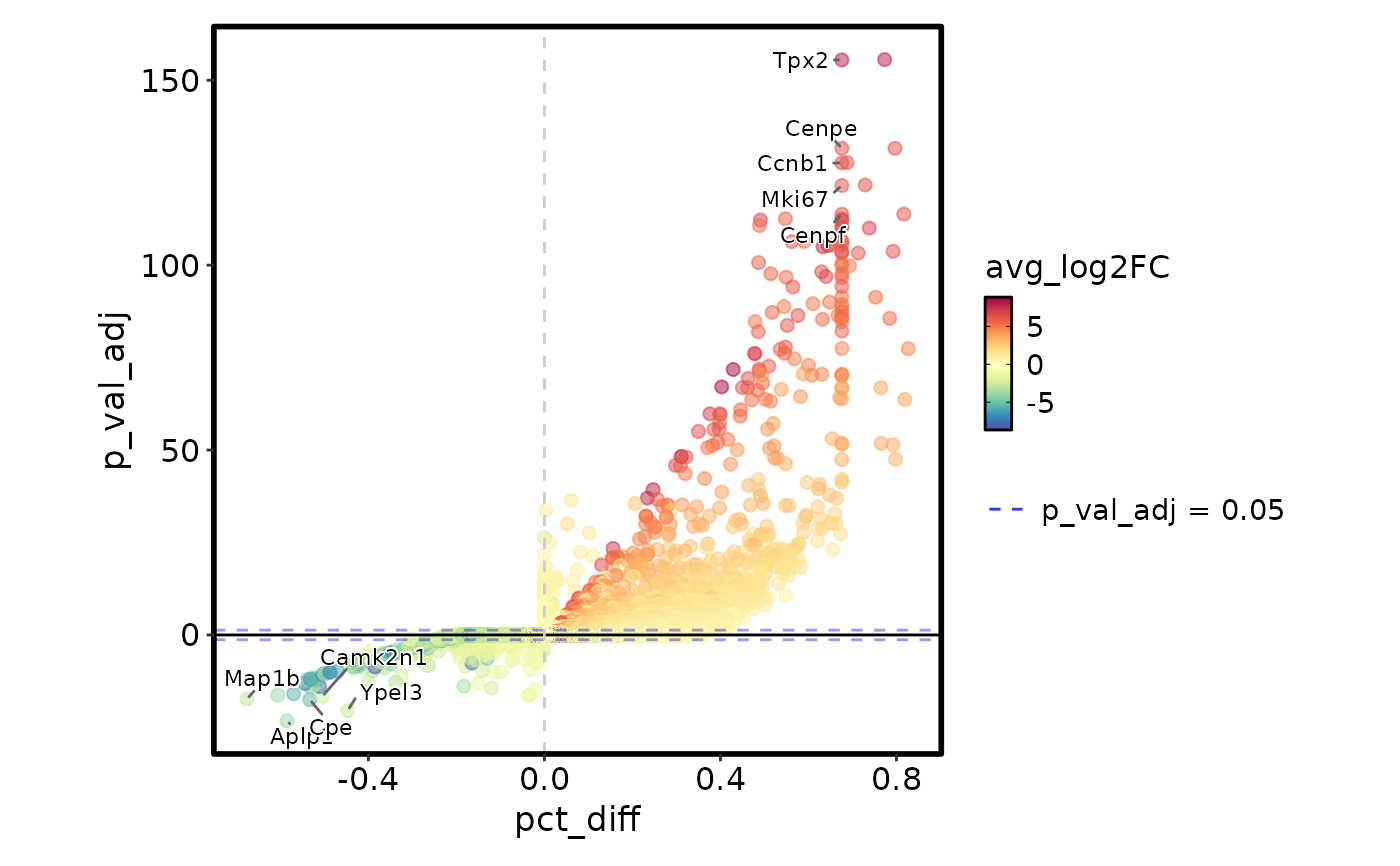 MarkersPlot(allmarkers, plot_type = "jitter", subset_by = "cluster")
MarkersPlot(allmarkers, plot_type = "jitter", subset_by = "cluster")
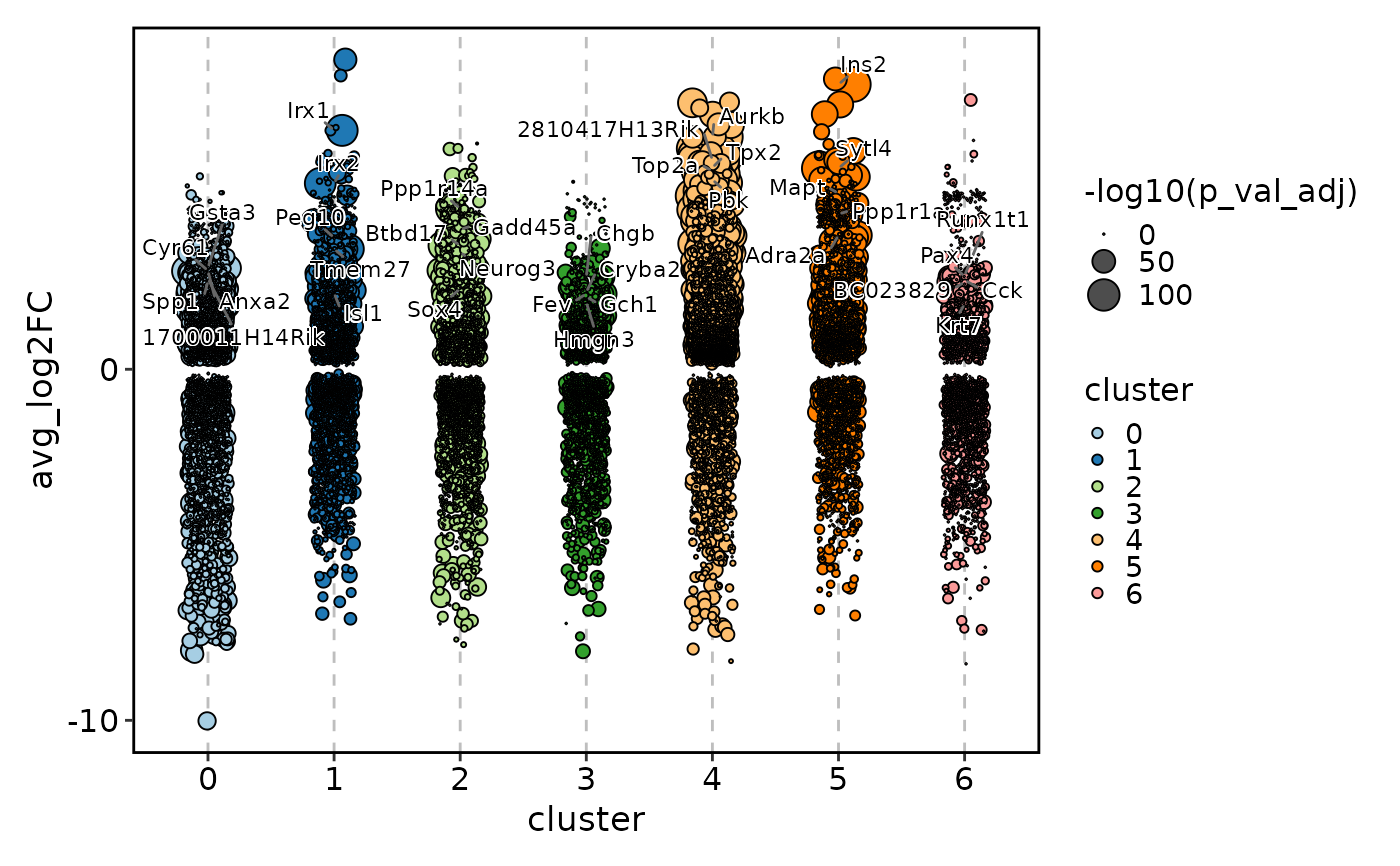 MarkersPlot(allmarkers, plot_type = "jitter_pct",
subset_by = "cluster", add_hline = 0, shape = 16)
MarkersPlot(allmarkers, plot_type = "jitter_pct",
subset_by = "cluster", add_hline = 0, shape = 16)
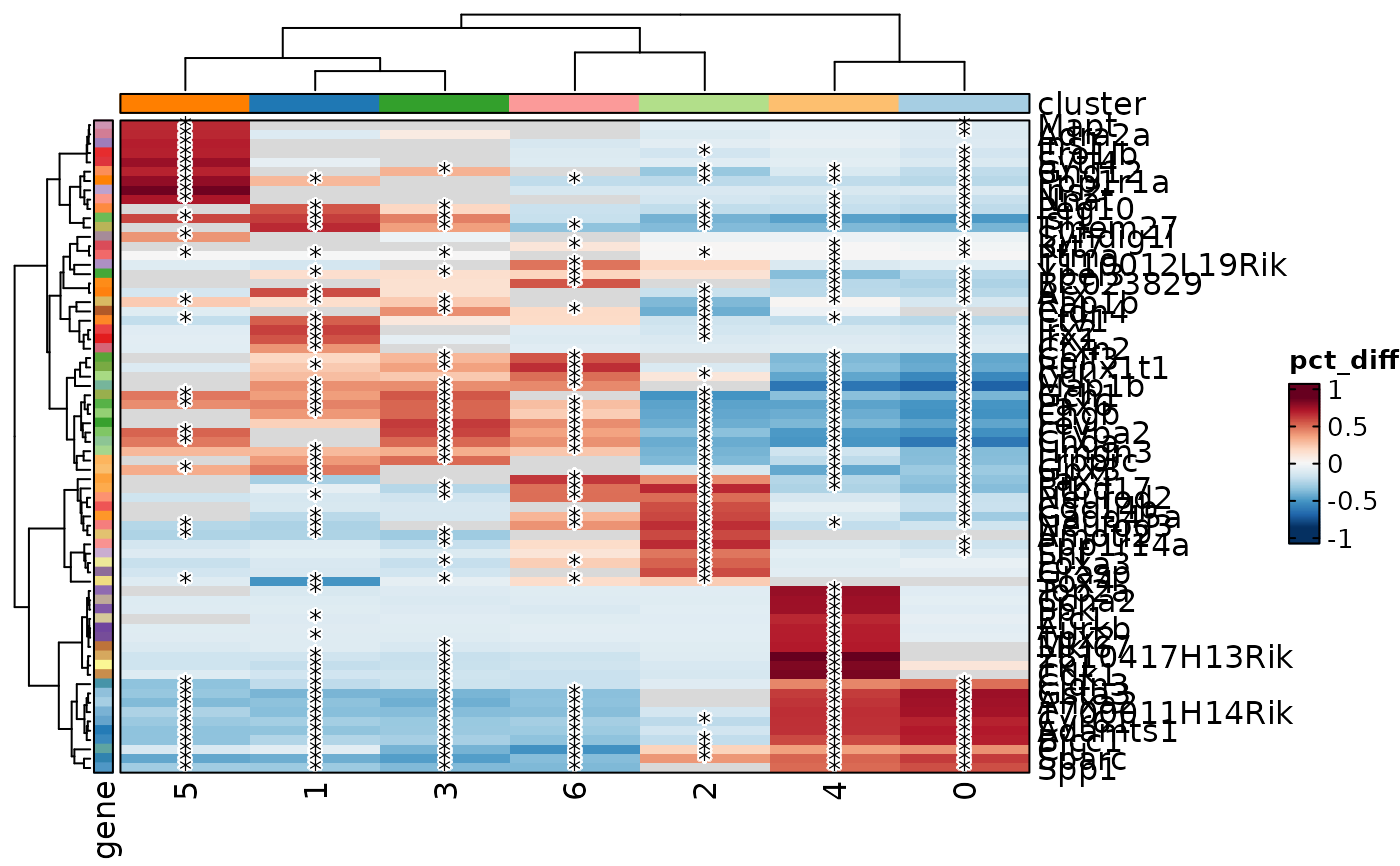
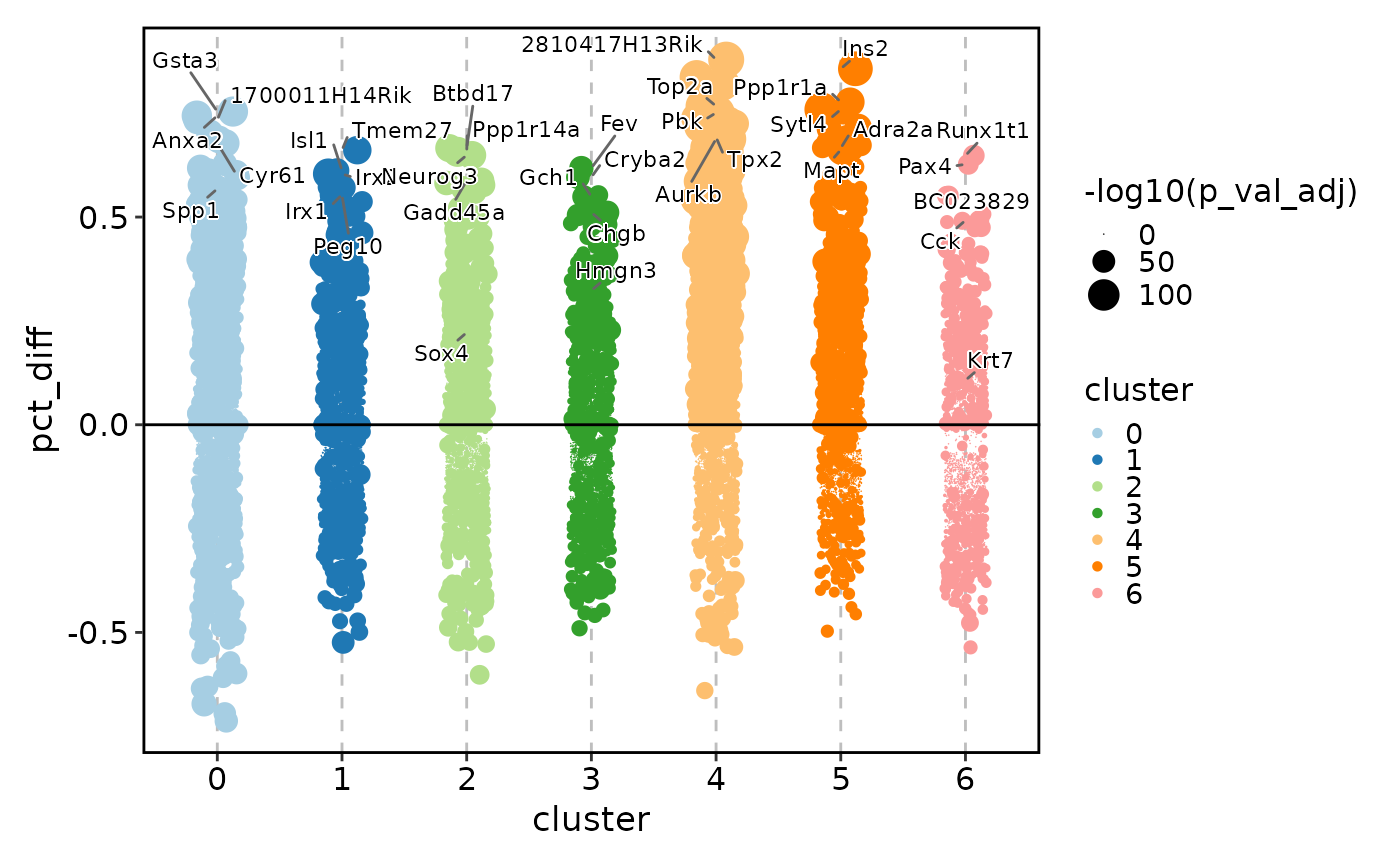 MarkersPlot(allmarkers, plot_type = "heatmap_log2fc", subset_by = "cluster")
MarkersPlot(allmarkers, plot_type = "heatmap_log2fc", subset_by = "cluster")
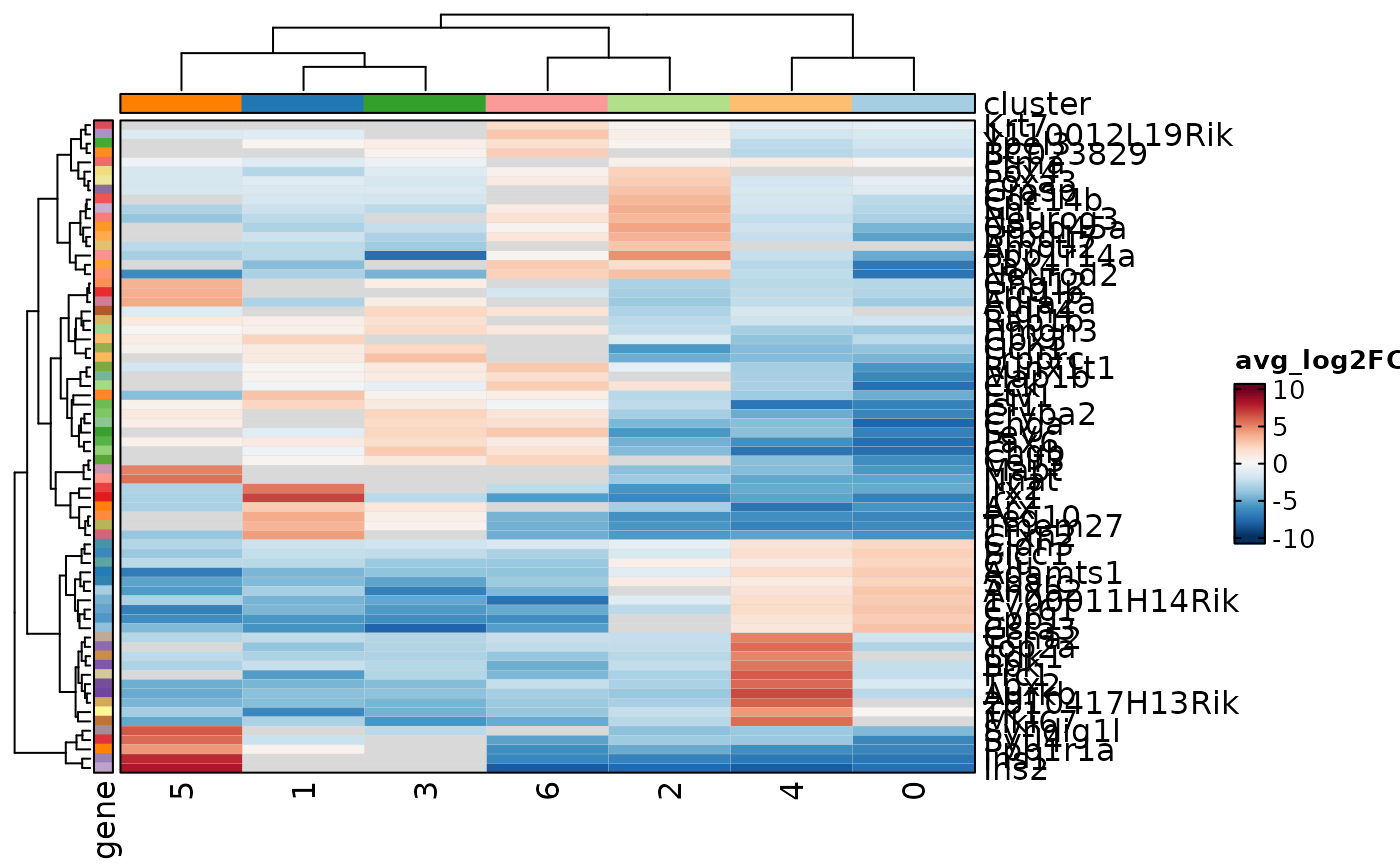 MarkersPlot(allmarkers, plot_type = "heatmap_pct", subset_by = "cluster",
cutoff = 0.05)
MarkersPlot(allmarkers, plot_type = "heatmap_pct", subset_by = "cluster",
cutoff = 0.05)
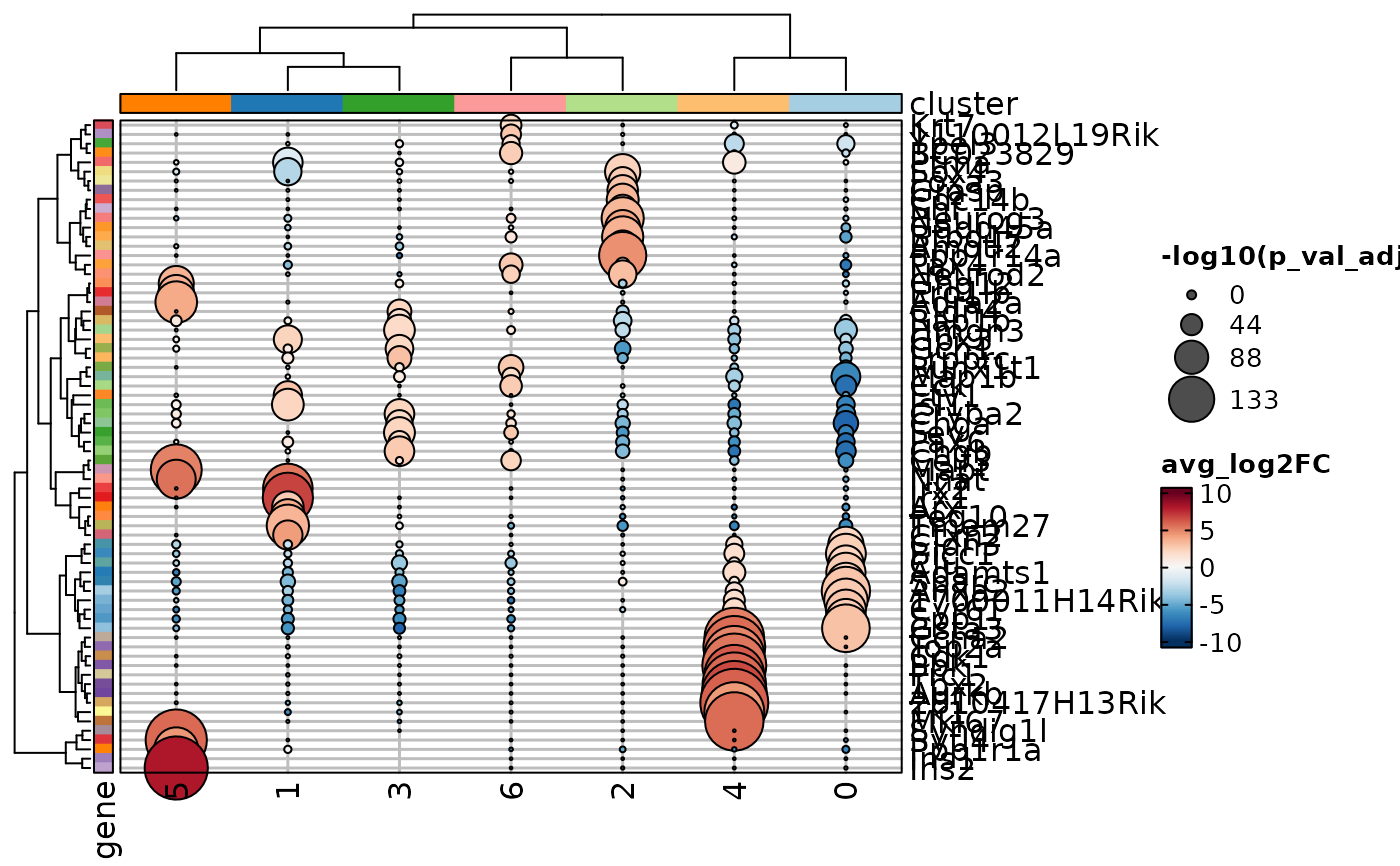
 MarkersPlot(allmarkers, plot_type = "dot_log2fc", subset_by = "cluster",
add_reticle = TRUE)
MarkersPlot(allmarkers, plot_type = "dot_log2fc", subset_by = "cluster",
add_reticle = TRUE)
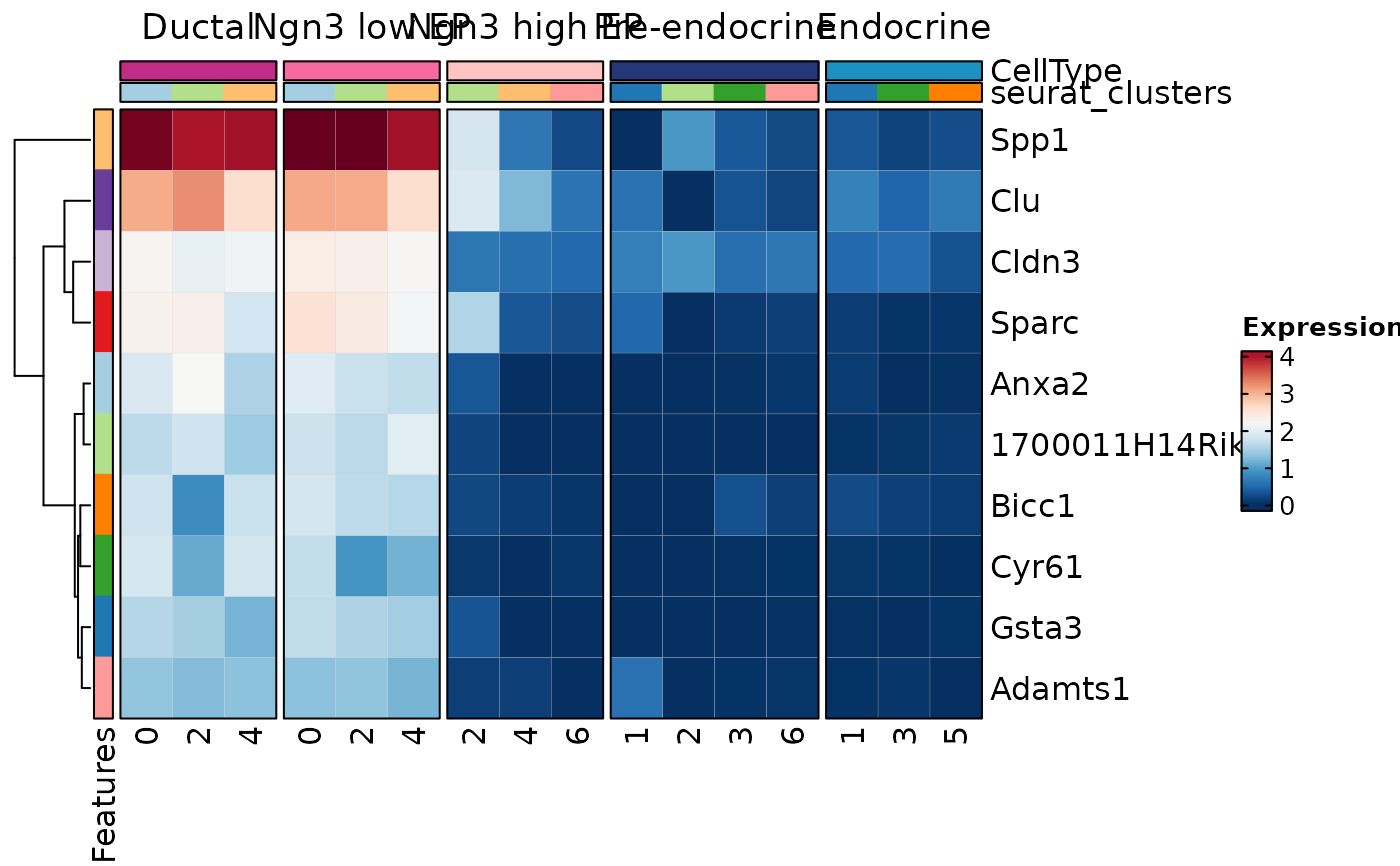
 MarkersPlot(allmarkers, object = pancreas_sub, plot_type = "heatmap",
columns_split_by = "CellType",
comparison_by = "cluster:seurat_clusters")
#> Warning: Layer counts isn't present in the assay object; returning NULL
MarkersPlot(allmarkers, object = pancreas_sub, plot_type = "heatmap",
columns_split_by = "CellType",
comparison_by = "cluster:seurat_clusters")
#> Warning: Layer counts isn't present in the assay object; returning NULL
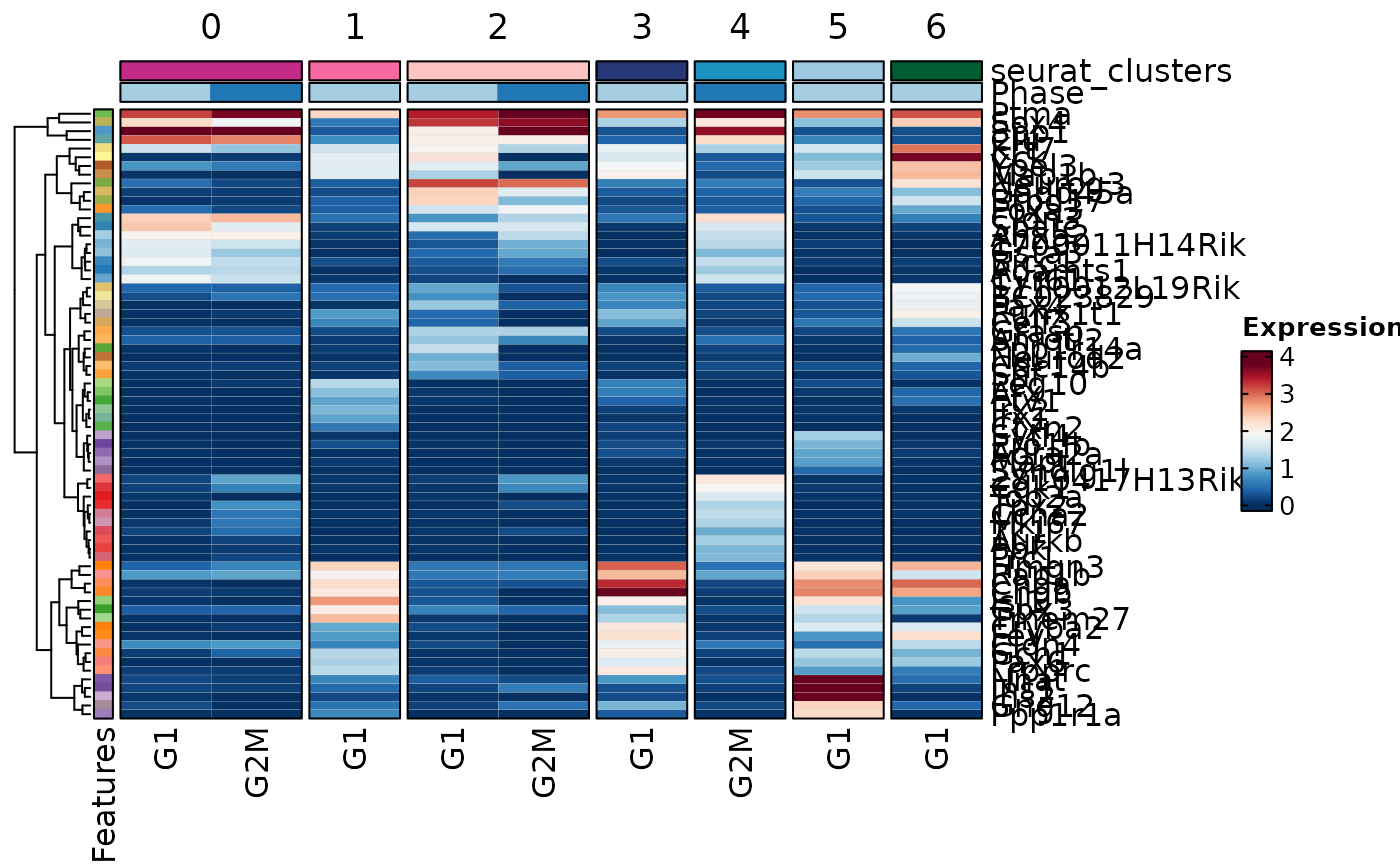
 # Suppose we did a DE between g1 and g2 in each cluster
allmarkers$comparison <- "g1:g2"
MarkersPlot(allmarkers, object = pancreas_sub, plot_type = "heatmap",
comparison_by = "Phase", subset_by = "cluster:seurat_clusters")
#> Warning: Layer counts isn't present in the assay object; returning NULL
# Suppose we did a DE between g1 and g2 in each cluster
allmarkers$comparison <- "g1:g2"
MarkersPlot(allmarkers, object = pancreas_sub, plot_type = "heatmap",
comparison_by = "Phase", subset_by = "cluster:seurat_clusters")
#> Warning: Layer counts isn't present in the assay object; returning NULL
 MarkersPlot(allmarkers, object = pancreas_sub, plot_type = "dot",
comparison_by = "Phase", subset_by = "cluster:seurat_clusters")
#> Warning: Layer counts isn't present in the assay object; returning NULL
MarkersPlot(allmarkers, object = pancreas_sub, plot_type = "dot",
comparison_by = "Phase", subset_by = "cluster:seurat_clusters")
#> Warning: Layer counts isn't present in the assay object; returning NULL
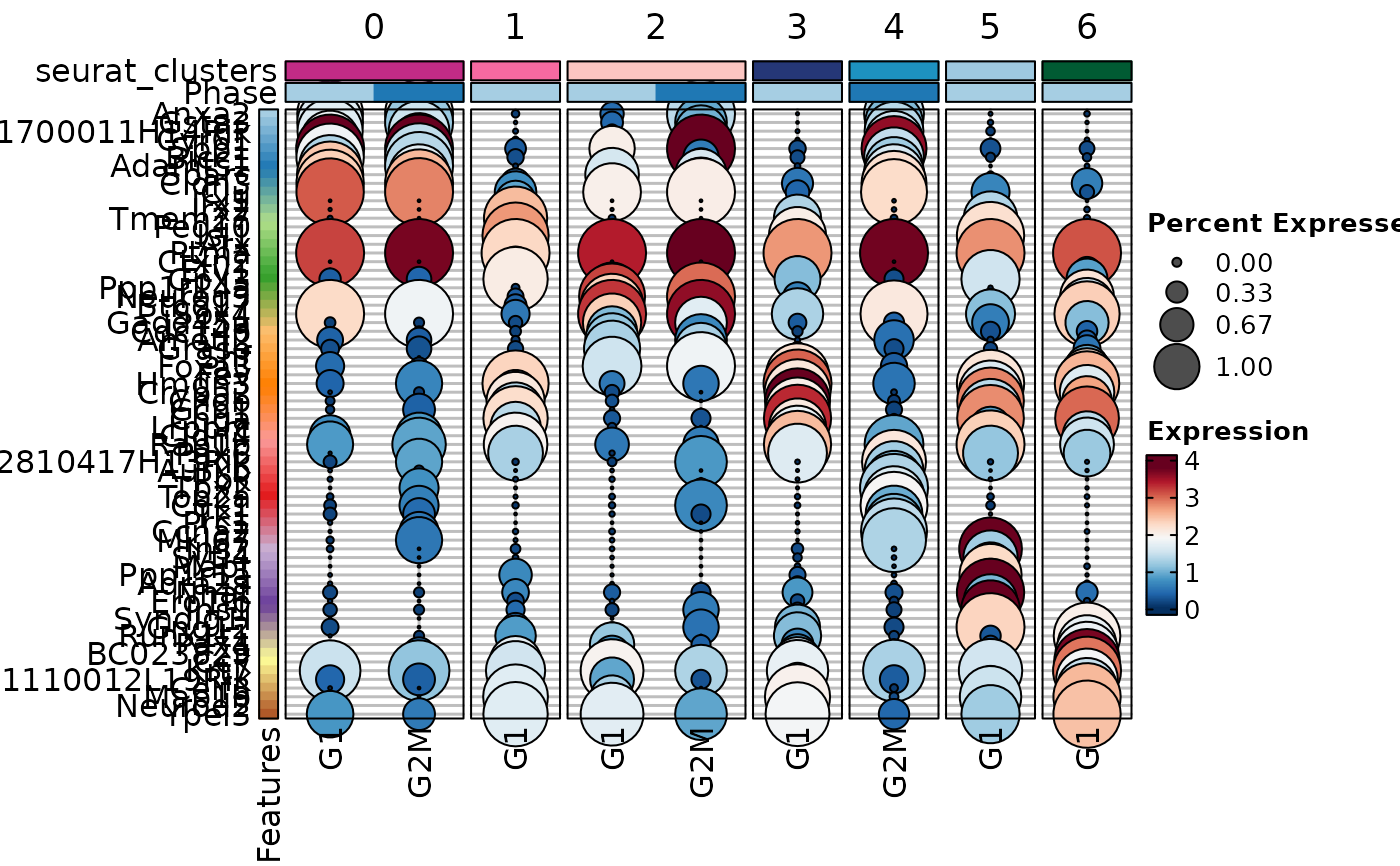 MarkersPlot(allmarkers, object = pancreas_sub, plot_type = "violin", select = 3,
comparison_by = "Phase", subset_by = "cluster:seurat_clusters")
#> Warning: Layer counts isn't present in the assay object; returning NULL
MarkersPlot(allmarkers, object = pancreas_sub, plot_type = "violin", select = 3,
comparison_by = "Phase", subset_by = "cluster:seurat_clusters")
#> Warning: Layer counts isn't present in the assay object; returning NULL
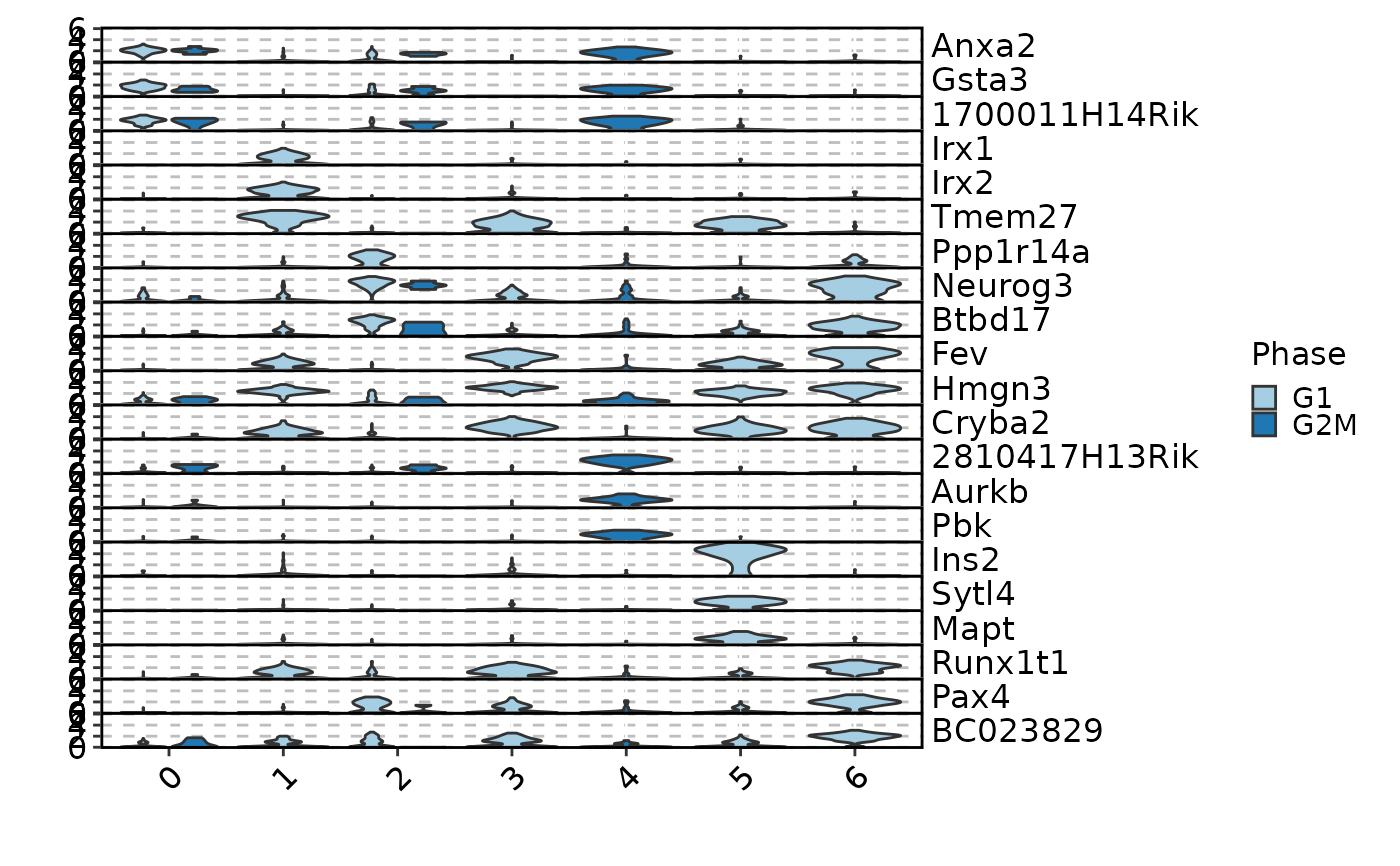 MarkersPlot(allmarkers, object = pancreas_sub, plot_type = "box", select = 3,
comparison_by = "Phase", subset_by = "cluster:seurat_clusters")
#> Warning: Layer counts isn't present in the assay object; returning NULL
MarkersPlot(allmarkers, object = pancreas_sub, plot_type = "box", select = 3,
comparison_by = "Phase", subset_by = "cluster:seurat_clusters")
#> Warning: Layer counts isn't present in the assay object; returning NULL
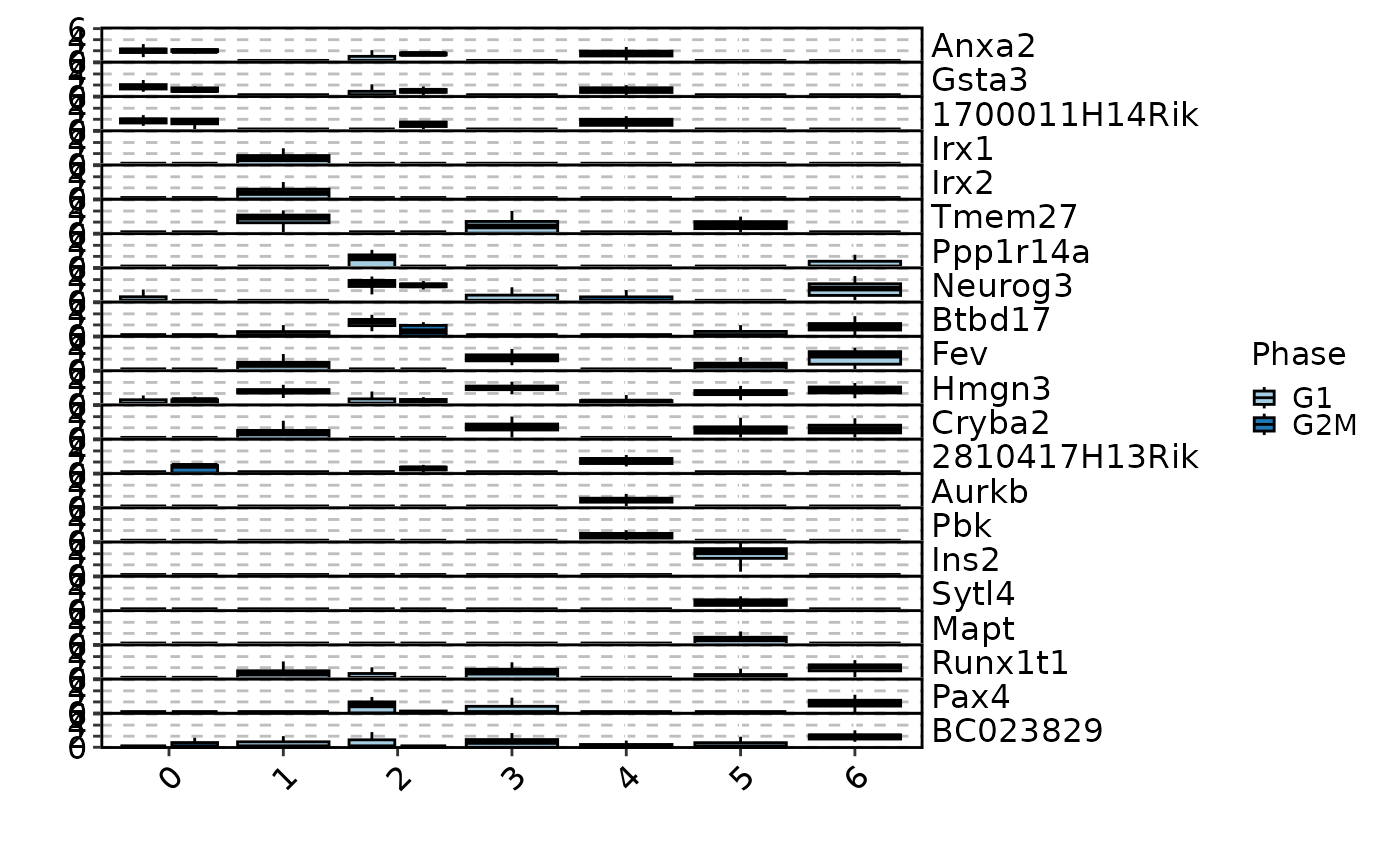 MarkersPlot(allmarkers, object = pancreas_sub, plot_type = "ridge", select = 2,
comparison_by = "Phase", subset_by = "cluster:seurat_clusters",
ncol = 2)
#> Warning: Layer counts isn't present in the assay object; returning NULL
#> Picking joint bandwidth of 0.155
#> Picking joint bandwidth of 0.222
#> Picking joint bandwidth of 0.464
#> Picking joint bandwidth of 0.307
#> Picking joint bandwidth of 0.32
#> Picking joint bandwidth of 0.222
#> Picking joint bandwidth of 0.322
#> Picking joint bandwidth of 0.246
#> Picking joint bandwidth of 0.265
#> Picking joint bandwidth of 0.153
#> Picking joint bandwidth of 0.326
#> Picking joint bandwidth of 0.464
#> Picking joint bandwidth of 0.094
#> Picking joint bandwidth of 0.306
#> Picking joint bandwidth of 0.116
#> Picking joint bandwidth of 0.0355
#> Picking joint bandwidth of 0.278
#> Picking joint bandwidth of 0.28
#> Picking joint bandwidth of 0.102
#> Picking joint bandwidth of 0.239
#> Picking joint bandwidth of 0.251
#> Picking joint bandwidth of 0.144
#> Picking joint bandwidth of 0.0735
#> Picking joint bandwidth of 0.031
#> Picking joint bandwidth of 0.202
#> Picking joint bandwidth of 0.0748
#> Picking joint bandwidth of 0.256
#> Picking joint bandwidth of 0.0755
#> Picking joint bandwidth of 0.201
#> Picking joint bandwidth of 0.234
#> Picking joint bandwidth of 0.512
#> Picking joint bandwidth of 0.351
#> Picking joint bandwidth of 0.515
#> Picking joint bandwidth of 0.267
#> Picking joint bandwidth of 0.381
#> Picking joint bandwidth of 0.351
#> Picking joint bandwidth of 0.174
#> Picking joint bandwidth of 0.361
#> Picking joint bandwidth of 0.368
#> Picking joint bandwidth of 0.362
#> Picking joint bandwidth of 0.461
#> Picking joint bandwidth of 0.259
#> Picking joint bandwidth of 0.031
#> Picking joint bandwidth of 0.327
#> Picking joint bandwidth of 0.0693
#> Picking joint bandwidth of 0.124
#> Picking joint bandwidth of 0.327
#> Picking joint bandwidth of 0.268
#> Picking joint bandwidth of 0.264
#> Picking joint bandwidth of 0.153
#> Picking joint bandwidth of 0.0669
#> Picking joint bandwidth of 0.0314
#> Picking joint bandwidth of 0.187
#> Picking joint bandwidth of 0.112
#> Picking joint bandwidth of 0.282
#> Picking joint bandwidth of 0.255
#> Picking joint bandwidth of 0.254
#> Picking joint bandwidth of 0.229
#> Picking joint bandwidth of 0.0263
#> Picking joint bandwidth of 0.0421
#> Picking joint bandwidth of 0.193
#> Picking joint bandwidth of 0.256
#> Picking joint bandwidth of 0.188
#> Picking joint bandwidth of 0.204
#> Picking joint bandwidth of 0.267
#> Picking joint bandwidth of 0.209
#> Picking joint bandwidth of 0.0543
#> Picking joint bandwidth of 0.0409
#> Picking joint bandwidth of 0.18
#> Picking joint bandwidth of 0.196
#> Picking joint bandwidth of 0.0491
#> Picking joint bandwidth of 0.0658
#> Picking joint bandwidth of 0.0579
#> Picking joint bandwidth of 0.0672
#> Picking joint bandwidth of 0.0904
#> Picking joint bandwidth of 0.185
#> Picking joint bandwidth of 0.241
#> Picking joint bandwidth of 0.195
#> Picking joint bandwidth of 0.0677
#> Picking joint bandwidth of 0.358
#> Picking joint bandwidth of 0.468
#> Picking joint bandwidth of 0.269
#> Picking joint bandwidth of 0.174
#> Picking joint bandwidth of 0.173
#> Picking joint bandwidth of 0.0847
#> Picking joint bandwidth of 0.0428
#> Picking joint bandwidth of 0.364
#> Picking joint bandwidth of 0.09
#> Picking joint bandwidth of 0.25
#> Picking joint bandwidth of 0.518
#> Picking joint bandwidth of 0.51
#> Picking joint bandwidth of 0.29
#> Picking joint bandwidth of 0.0588
#> Picking joint bandwidth of 0.0403
#> Picking joint bandwidth of 0.0428
#> Picking joint bandwidth of 0.364
#> Picking joint bandwidth of 0.259
#> Picking joint bandwidth of 0.285
#> Picking joint bandwidth of 0.155
#> Picking joint bandwidth of 0.222
#> Picking joint bandwidth of 0.464
#> Picking joint bandwidth of 0.307
#> Picking joint bandwidth of 0.32
#> Picking joint bandwidth of 0.222
#> Picking joint bandwidth of 0.322
#> Picking joint bandwidth of 0.246
#> Picking joint bandwidth of 0.265
#> Picking joint bandwidth of 0.153
#> Picking joint bandwidth of 0.326
#> Picking joint bandwidth of 0.464
#> Picking joint bandwidth of 0.094
#> Picking joint bandwidth of 0.306
#> Picking joint bandwidth of 0.116
#> Picking joint bandwidth of 0.0355
#> Picking joint bandwidth of 0.278
#> Picking joint bandwidth of 0.28
#> Picking joint bandwidth of 0.102
#> Picking joint bandwidth of 0.239
#> Picking joint bandwidth of 0.251
#> Picking joint bandwidth of 0.144
#> Picking joint bandwidth of 0.0735
#> Picking joint bandwidth of 0.031
#> Picking joint bandwidth of 0.202
#> Picking joint bandwidth of 0.0748
#> Picking joint bandwidth of 0.256
#> Picking joint bandwidth of 0.0755
#> Picking joint bandwidth of 0.201
#> Picking joint bandwidth of 0.234
#> Picking joint bandwidth of 0.512
#> Picking joint bandwidth of 0.351
#> Picking joint bandwidth of 0.515
#> Picking joint bandwidth of 0.267
#> Picking joint bandwidth of 0.381
#> Picking joint bandwidth of 0.351
#> Picking joint bandwidth of 0.174
#> Picking joint bandwidth of 0.361
#> Picking joint bandwidth of 0.368
#> Picking joint bandwidth of 0.362
#> Picking joint bandwidth of 0.461
#> Picking joint bandwidth of 0.259
#> Picking joint bandwidth of 0.031
#> Picking joint bandwidth of 0.327
#> Picking joint bandwidth of 0.0693
#> Picking joint bandwidth of 0.124
#> Picking joint bandwidth of 0.327
#> Picking joint bandwidth of 0.268
#> Picking joint bandwidth of 0.264
#> Picking joint bandwidth of 0.153
#> Picking joint bandwidth of 0.0669
#> Picking joint bandwidth of 0.0314
#> Picking joint bandwidth of 0.187
#> Picking joint bandwidth of 0.112
#> Picking joint bandwidth of 0.282
#> Picking joint bandwidth of 0.255
#> Picking joint bandwidth of 0.254
#> Picking joint bandwidth of 0.229
#> Picking joint bandwidth of 0.0263
#> Picking joint bandwidth of 0.0421
#> Picking joint bandwidth of 0.193
#> Picking joint bandwidth of 0.256
#> Picking joint bandwidth of 0.188
#> Picking joint bandwidth of 0.204
#> Picking joint bandwidth of 0.267
#> Picking joint bandwidth of 0.209
#> Picking joint bandwidth of 0.0543
#> Picking joint bandwidth of 0.0409
#> Picking joint bandwidth of 0.18
#> Picking joint bandwidth of 0.196
#> Picking joint bandwidth of 0.0491
#> Picking joint bandwidth of 0.0658
#> Picking joint bandwidth of 0.0579
#> Picking joint bandwidth of 0.0672
#> Picking joint bandwidth of 0.0904
#> Picking joint bandwidth of 0.185
#> Picking joint bandwidth of 0.241
#> Picking joint bandwidth of 0.195
#> Picking joint bandwidth of 0.0677
#> Picking joint bandwidth of 0.358
#> Picking joint bandwidth of 0.468
#> Picking joint bandwidth of 0.269
#> Picking joint bandwidth of 0.174
#> Picking joint bandwidth of 0.173
#> Picking joint bandwidth of 0.0847
#> Picking joint bandwidth of 0.0428
#> Picking joint bandwidth of 0.364
#> Picking joint bandwidth of 0.09
#> Picking joint bandwidth of 0.25
#> Picking joint bandwidth of 0.518
#> Picking joint bandwidth of 0.51
#> Picking joint bandwidth of 0.29
#> Picking joint bandwidth of 0.0588
#> Picking joint bandwidth of 0.0403
#> Picking joint bandwidth of 0.0428
#> Picking joint bandwidth of 0.364
#> Picking joint bandwidth of 0.259
#> Picking joint bandwidth of 0.285
MarkersPlot(allmarkers, object = pancreas_sub, plot_type = "ridge", select = 2,
comparison_by = "Phase", subset_by = "cluster:seurat_clusters",
ncol = 2)
#> Warning: Layer counts isn't present in the assay object; returning NULL
#> Picking joint bandwidth of 0.155
#> Picking joint bandwidth of 0.222
#> Picking joint bandwidth of 0.464
#> Picking joint bandwidth of 0.307
#> Picking joint bandwidth of 0.32
#> Picking joint bandwidth of 0.222
#> Picking joint bandwidth of 0.322
#> Picking joint bandwidth of 0.246
#> Picking joint bandwidth of 0.265
#> Picking joint bandwidth of 0.153
#> Picking joint bandwidth of 0.326
#> Picking joint bandwidth of 0.464
#> Picking joint bandwidth of 0.094
#> Picking joint bandwidth of 0.306
#> Picking joint bandwidth of 0.116
#> Picking joint bandwidth of 0.0355
#> Picking joint bandwidth of 0.278
#> Picking joint bandwidth of 0.28
#> Picking joint bandwidth of 0.102
#> Picking joint bandwidth of 0.239
#> Picking joint bandwidth of 0.251
#> Picking joint bandwidth of 0.144
#> Picking joint bandwidth of 0.0735
#> Picking joint bandwidth of 0.031
#> Picking joint bandwidth of 0.202
#> Picking joint bandwidth of 0.0748
#> Picking joint bandwidth of 0.256
#> Picking joint bandwidth of 0.0755
#> Picking joint bandwidth of 0.201
#> Picking joint bandwidth of 0.234
#> Picking joint bandwidth of 0.512
#> Picking joint bandwidth of 0.351
#> Picking joint bandwidth of 0.515
#> Picking joint bandwidth of 0.267
#> Picking joint bandwidth of 0.381
#> Picking joint bandwidth of 0.351
#> Picking joint bandwidth of 0.174
#> Picking joint bandwidth of 0.361
#> Picking joint bandwidth of 0.368
#> Picking joint bandwidth of 0.362
#> Picking joint bandwidth of 0.461
#> Picking joint bandwidth of 0.259
#> Picking joint bandwidth of 0.031
#> Picking joint bandwidth of 0.327
#> Picking joint bandwidth of 0.0693
#> Picking joint bandwidth of 0.124
#> Picking joint bandwidth of 0.327
#> Picking joint bandwidth of 0.268
#> Picking joint bandwidth of 0.264
#> Picking joint bandwidth of 0.153
#> Picking joint bandwidth of 0.0669
#> Picking joint bandwidth of 0.0314
#> Picking joint bandwidth of 0.187
#> Picking joint bandwidth of 0.112
#> Picking joint bandwidth of 0.282
#> Picking joint bandwidth of 0.255
#> Picking joint bandwidth of 0.254
#> Picking joint bandwidth of 0.229
#> Picking joint bandwidth of 0.0263
#> Picking joint bandwidth of 0.0421
#> Picking joint bandwidth of 0.193
#> Picking joint bandwidth of 0.256
#> Picking joint bandwidth of 0.188
#> Picking joint bandwidth of 0.204
#> Picking joint bandwidth of 0.267
#> Picking joint bandwidth of 0.209
#> Picking joint bandwidth of 0.0543
#> Picking joint bandwidth of 0.0409
#> Picking joint bandwidth of 0.18
#> Picking joint bandwidth of 0.196
#> Picking joint bandwidth of 0.0491
#> Picking joint bandwidth of 0.0658
#> Picking joint bandwidth of 0.0579
#> Picking joint bandwidth of 0.0672
#> Picking joint bandwidth of 0.0904
#> Picking joint bandwidth of 0.185
#> Picking joint bandwidth of 0.241
#> Picking joint bandwidth of 0.195
#> Picking joint bandwidth of 0.0677
#> Picking joint bandwidth of 0.358
#> Picking joint bandwidth of 0.468
#> Picking joint bandwidth of 0.269
#> Picking joint bandwidth of 0.174
#> Picking joint bandwidth of 0.173
#> Picking joint bandwidth of 0.0847
#> Picking joint bandwidth of 0.0428
#> Picking joint bandwidth of 0.364
#> Picking joint bandwidth of 0.09
#> Picking joint bandwidth of 0.25
#> Picking joint bandwidth of 0.518
#> Picking joint bandwidth of 0.51
#> Picking joint bandwidth of 0.29
#> Picking joint bandwidth of 0.0588
#> Picking joint bandwidth of 0.0403
#> Picking joint bandwidth of 0.0428
#> Picking joint bandwidth of 0.364
#> Picking joint bandwidth of 0.259
#> Picking joint bandwidth of 0.285
#> Picking joint bandwidth of 0.155
#> Picking joint bandwidth of 0.222
#> Picking joint bandwidth of 0.464
#> Picking joint bandwidth of 0.307
#> Picking joint bandwidth of 0.32
#> Picking joint bandwidth of 0.222
#> Picking joint bandwidth of 0.322
#> Picking joint bandwidth of 0.246
#> Picking joint bandwidth of 0.265
#> Picking joint bandwidth of 0.153
#> Picking joint bandwidth of 0.326
#> Picking joint bandwidth of 0.464
#> Picking joint bandwidth of 0.094
#> Picking joint bandwidth of 0.306
#> Picking joint bandwidth of 0.116
#> Picking joint bandwidth of 0.0355
#> Picking joint bandwidth of 0.278
#> Picking joint bandwidth of 0.28
#> Picking joint bandwidth of 0.102
#> Picking joint bandwidth of 0.239
#> Picking joint bandwidth of 0.251
#> Picking joint bandwidth of 0.144
#> Picking joint bandwidth of 0.0735
#> Picking joint bandwidth of 0.031
#> Picking joint bandwidth of 0.202
#> Picking joint bandwidth of 0.0748
#> Picking joint bandwidth of 0.256
#> Picking joint bandwidth of 0.0755
#> Picking joint bandwidth of 0.201
#> Picking joint bandwidth of 0.234
#> Picking joint bandwidth of 0.512
#> Picking joint bandwidth of 0.351
#> Picking joint bandwidth of 0.515
#> Picking joint bandwidth of 0.267
#> Picking joint bandwidth of 0.381
#> Picking joint bandwidth of 0.351
#> Picking joint bandwidth of 0.174
#> Picking joint bandwidth of 0.361
#> Picking joint bandwidth of 0.368
#> Picking joint bandwidth of 0.362
#> Picking joint bandwidth of 0.461
#> Picking joint bandwidth of 0.259
#> Picking joint bandwidth of 0.031
#> Picking joint bandwidth of 0.327
#> Picking joint bandwidth of 0.0693
#> Picking joint bandwidth of 0.124
#> Picking joint bandwidth of 0.327
#> Picking joint bandwidth of 0.268
#> Picking joint bandwidth of 0.264
#> Picking joint bandwidth of 0.153
#> Picking joint bandwidth of 0.0669
#> Picking joint bandwidth of 0.0314
#> Picking joint bandwidth of 0.187
#> Picking joint bandwidth of 0.112
#> Picking joint bandwidth of 0.282
#> Picking joint bandwidth of 0.255
#> Picking joint bandwidth of 0.254
#> Picking joint bandwidth of 0.229
#> Picking joint bandwidth of 0.0263
#> Picking joint bandwidth of 0.0421
#> Picking joint bandwidth of 0.193
#> Picking joint bandwidth of 0.256
#> Picking joint bandwidth of 0.188
#> Picking joint bandwidth of 0.204
#> Picking joint bandwidth of 0.267
#> Picking joint bandwidth of 0.209
#> Picking joint bandwidth of 0.0543
#> Picking joint bandwidth of 0.0409
#> Picking joint bandwidth of 0.18
#> Picking joint bandwidth of 0.196
#> Picking joint bandwidth of 0.0491
#> Picking joint bandwidth of 0.0658
#> Picking joint bandwidth of 0.0579
#> Picking joint bandwidth of 0.0672
#> Picking joint bandwidth of 0.0904
#> Picking joint bandwidth of 0.185
#> Picking joint bandwidth of 0.241
#> Picking joint bandwidth of 0.195
#> Picking joint bandwidth of 0.0677
#> Picking joint bandwidth of 0.358
#> Picking joint bandwidth of 0.468
#> Picking joint bandwidth of 0.269
#> Picking joint bandwidth of 0.174
#> Picking joint bandwidth of 0.173
#> Picking joint bandwidth of 0.0847
#> Picking joint bandwidth of 0.0428
#> Picking joint bandwidth of 0.364
#> Picking joint bandwidth of 0.09
#> Picking joint bandwidth of 0.25
#> Picking joint bandwidth of 0.518
#> Picking joint bandwidth of 0.51
#> Picking joint bandwidth of 0.29
#> Picking joint bandwidth of 0.0588
#> Picking joint bandwidth of 0.0403
#> Picking joint bandwidth of 0.0428
#> Picking joint bandwidth of 0.364
#> Picking joint bandwidth of 0.259
#> Picking joint bandwidth of 0.285
 # }
# }