Visualize the statistics of the clones.
Usage
ClonalStatPlot(
data,
clones = NULL,
top = NULL,
orderby = NULL,
clone_call = "aa",
chain = "both",
plot_type = c("bar", "box", "violin", "heatmap", "pies", "sankey", "alluvial", "trend"),
group_by = "Sample",
groups = NULL,
subgroup_by = NULL,
subgroups = NULL,
within_subgroup = match.arg(plot_type) != "pies",
relabel = FALSE,
facet_by = NULL,
split_by = NULL,
y = NULL,
xlab = NULL,
ylab = NULL,
...
)Arguments
- data
The product of scRepertoire::combineTCR, scRepertoire::combineTCR, or scRepertoire::combineExpression.
- clones
The specific clones to track. This argument must be provided. If multiple character values are provided, they will be treated as clone IDs. If a single character value is provided with parentheses, it will be evaluated as an expression to select the clones. The clones will be selected per subgrouping/facetting/splitting group. For example, if you have
top(3)will select the top 3 clones in each facetting/splitting group. You can change this behavior by passing thegroupsargument explicitly. For exampletop(3, groups = "Sample")will select the top 3 clones in each sample. For expression, see alsoclone_selectors. This can also be a named list of expressions, which need to be quoted. Then basic unit for visualization will be the the clone groups defined by the names of the list, instead of single clones.- top
The number of top clones to select. Default is 10. A shortcut for
top(10)ifclonesis not provided. Ifclonesis provided, this will be a limit for the number of clones selected (based on theorderbyexpression). Ifclonesis a list, this will be applied to each clone group.- orderby
An expression to order the clones by. Default is NULL. Note that the clones will be ordered by the value of this expression in descending order.
- clone_call
How to call the clone - VDJC gene (gene), CDR3 nucleotide (nt), CDR3 amino acid (aa), VDJC gene + CDR3 nucleotide (strict) or a custom variable in the data
- chain
indicate if both or a specific chain should be used - e.g. "both", "TRA", "TRG", "IGH", "IGL"
- plot_type
The type of plot to use. Default is "bar". Possible values are "trend", "sankey", and "alluvial" (alias of "sankey").
- group_by
The column name in the meta data to group the cells. Default: "Sample"
- groups
The groups to include in the plot. Default is NULL. If NULL, all the groups in
group_bywill be included.- subgroup_by
The column name in the meta data to subgroup the nodes (group nodes on each
x). Default: NULL. This argument is only supported for "sankey"/"alluvial" plot. If NULL, the nodes will be grouped/colored by the clones- subgroups
The subgroups to include in the plot. Default is NULL.
- within_subgroup
Whether to select the clones within each subgroup.
- relabel
Whether to relabel the clones. Default is FALSE. The clone ids, especially using CDR3 sequences, can be long and hard to read. If TRUE, the clones will be relabeled as "clone1", "clone2", etc. Only works for visualizations for single clones.
- facet_by
The column name in the meta data to facet the plots. Default: NULL. This argument is not supported and will raise an error if provided.
- split_by
The column name in the meta data to split the plots. Default: NULL
- y
The y-axis variable to use for the plot. Default is NULL.
For
barplot, Either "TotalSize" or "Count" can be used, representing the total size (# cells) of the selected clones or the number of selected clones, respectively.
- xlab
The x-axis label. Default is NULL.
- ylab
The y-axis label. Default is NULL.
- ...
Other arguments passed to the specific plot function.
For
barplot, seeplotthis::BarPlot().For
trendplot, seeplotthis::TrendPlot().For
sankeyplot, seeplotthis::SankeyPlot().
Examples
# \donttest{
set.seed(8525)
data(contig_list, package = "scRepertoire")
data <- scRepertoire::combineTCR(contig_list,
samples = c("P17B", "P17L", "P18B", "P18L", "P19B","P19L", "P20B", "P20L"))
data <- scRepertoire::addVariable(data,
variable.name = "Type",
variables = rep(c("B", "L"), 4)
)
data <- scRepertoire::addVariable(data,
variable.name = "Subject",
variables = rep(c("P17", "P18", "P19", "P20"), each = 2)
)
# add a fake variable (e.g. cell type from scRNA-seq)
data <- lapply(data, function(x) {
x$CellType <- sample(c("CD4", "CD8", "B", "NK"), nrow(x), replace = TRUE)
# x <- x[x$CTaa == "CAVRKTTGTASKLTF_CASSLFGDKGETQYF", , drop = F]
return(x)
})
# showing the top 10 clones in P17B and P17L
ClonalStatPlot(data, group_by = "Sample", groups = c("P17B", "P17L"))
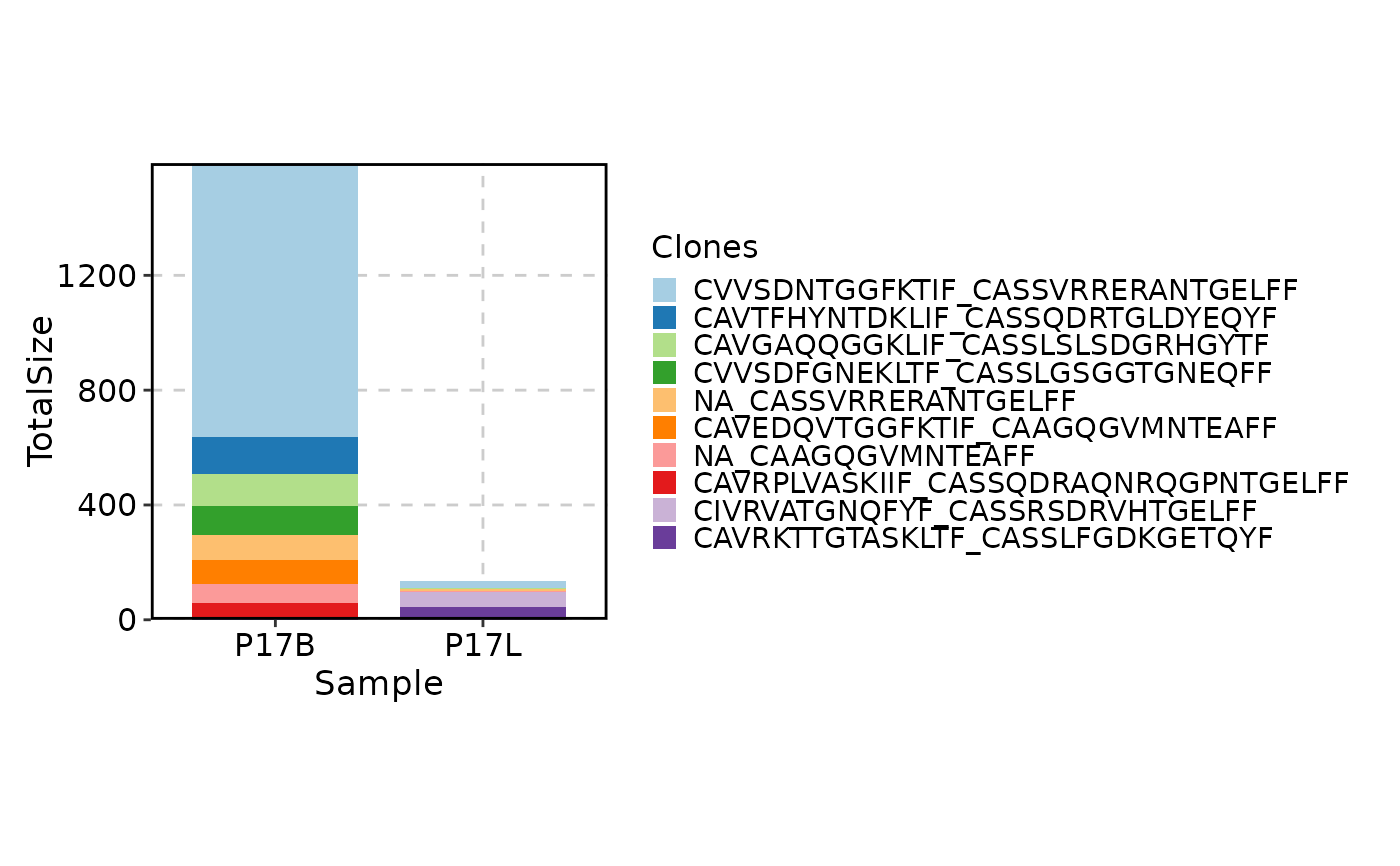 # showing the top 10 clones in P17B and P17L, with the clones relabeled
ClonalStatPlot(data, group_by = "Sample", groups = c("P17B", "P17L"), relabel = TRUE)
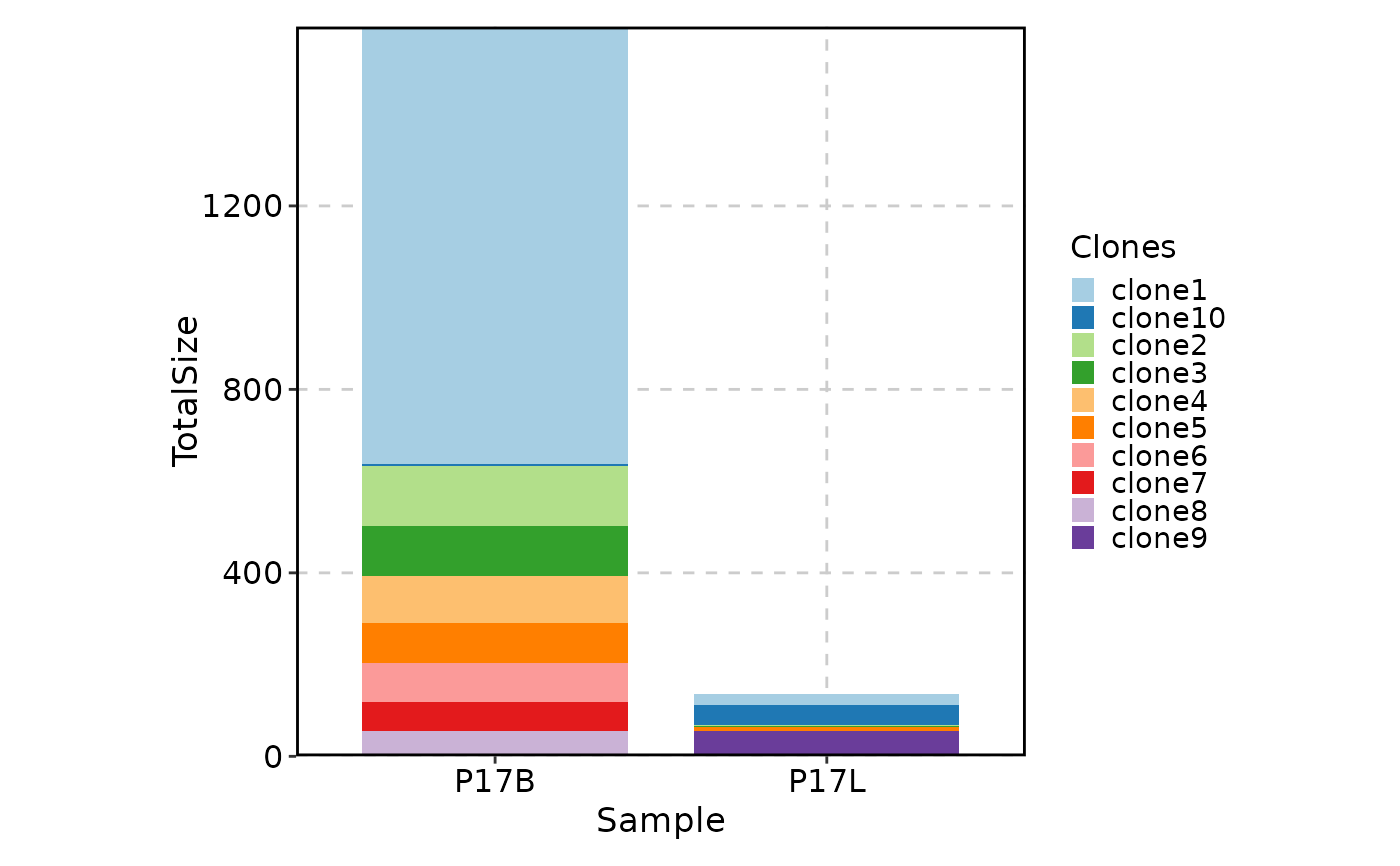
# showing the top 10 clones in P17B and P17L, with the clones relabeled
ClonalStatPlot(data, group_by = "Sample", groups = c("P17B", "P17L"), relabel = TRUE)
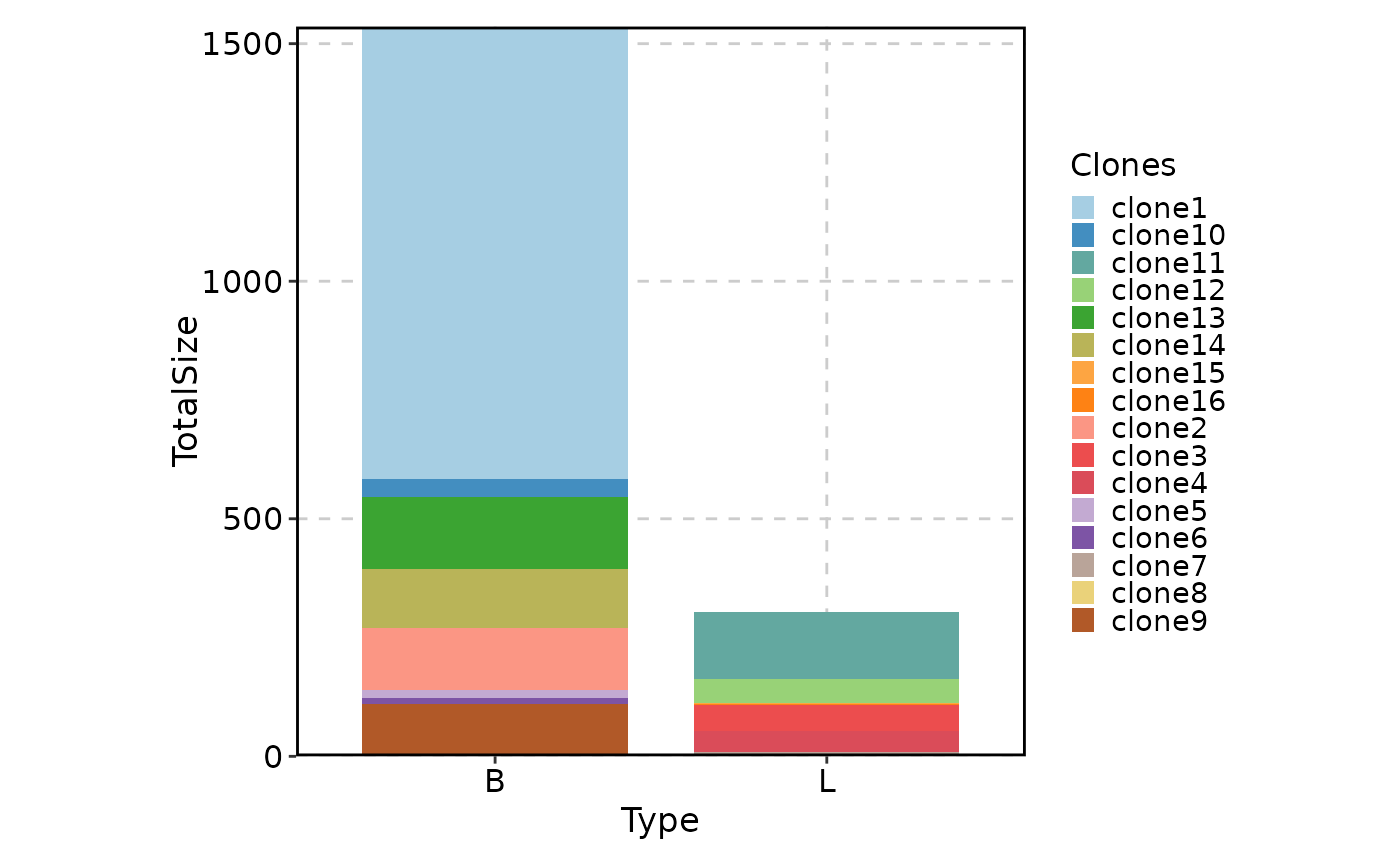
 # showing the top 2 clones in groups B and L, with subgroups in each group
ClonalStatPlot(data, group_by = "Type", subgroup_by = "Sample", top = 2,
subgroups = c("P17B", "P17L", "P18B", "P18L", "P19B","P19L"), relabel = TRUE)
# showing the top 2 clones in groups B and L, with subgroups in each group
ClonalStatPlot(data, group_by = "Type", subgroup_by = "Sample", top = 2,
subgroups = c("P17B", "P17L", "P18B", "P18L", "P19B","P19L"), relabel = TRUE)
 # showing selected clones in P17B and P17L
ClonalStatPlot(data, group_by = "Sample", groups = c("P17B", "P17L"),
clones = c("CVVSDNTGGFKTIF_CASSVRRERANTGELFF", "NA_CASSVRRERANTGELFF"), relabel = TRUE)
# showing selected clones in P17B and P17L
ClonalStatPlot(data, group_by = "Sample", groups = c("P17B", "P17L"),
clones = c("CVVSDNTGGFKTIF_CASSVRRERANTGELFF", "NA_CASSVRRERANTGELFF"), relabel = TRUE)
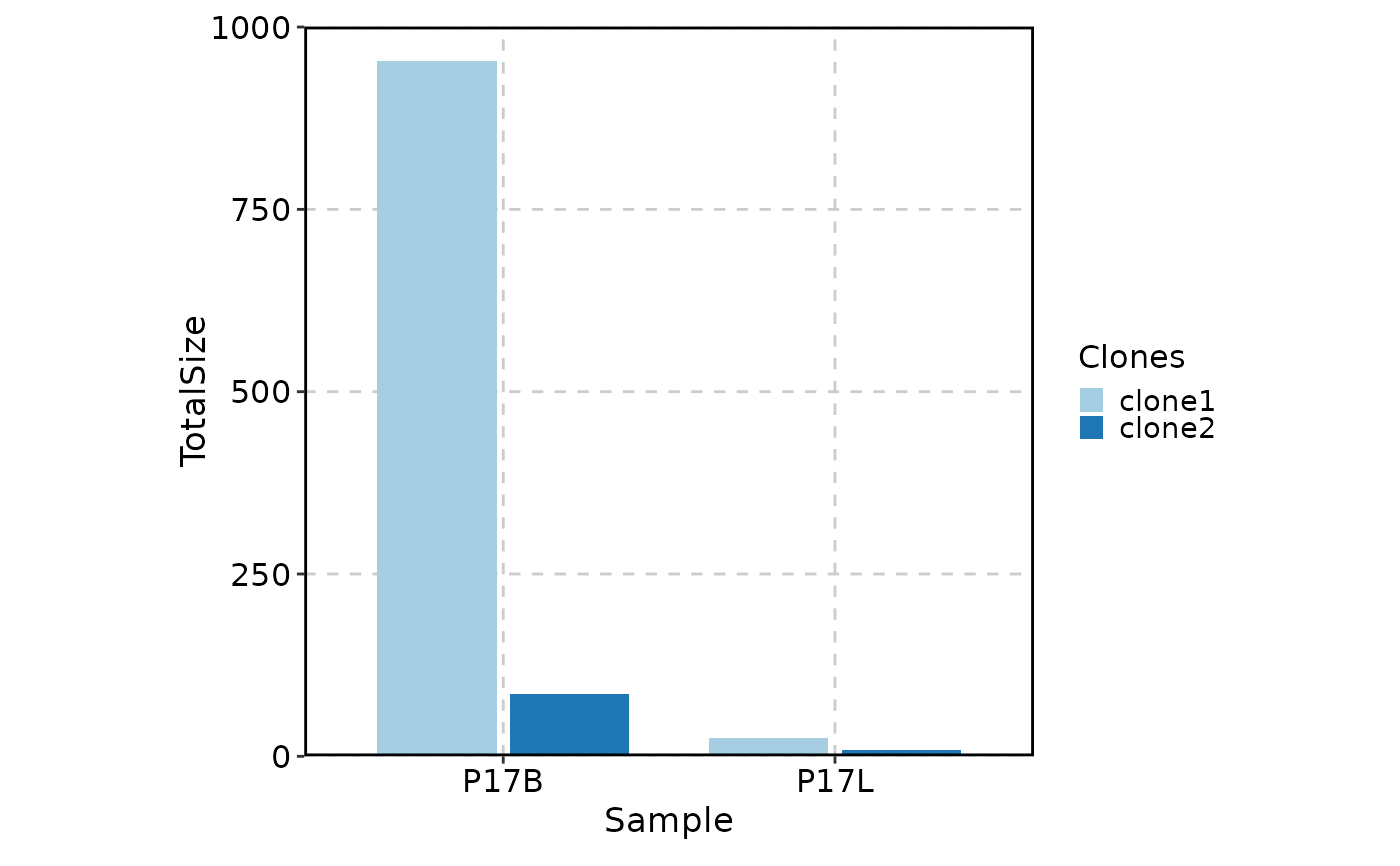 # facetting is supported
ClonalStatPlot(data, group_by = "Subject", groups = c("P17", "P19"),
facet_by = "Type", relabel = TRUE)
# facetting is supported
ClonalStatPlot(data, group_by = "Subject", groups = c("P17", "P19"),
facet_by = "Type", relabel = TRUE)
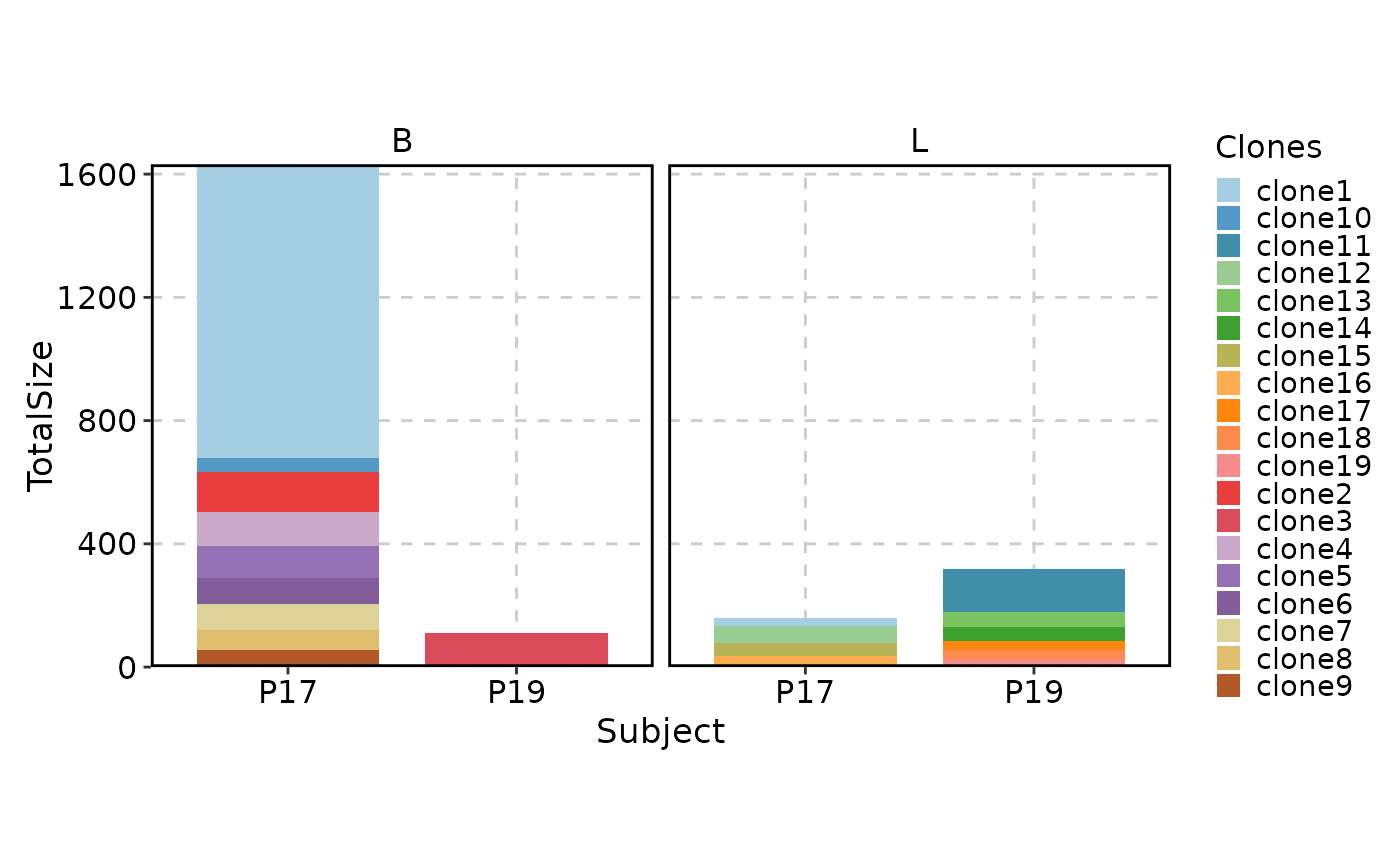 # as well as splitting
ClonalStatPlot(data, group_by = "Subject", groups = c("P17", "P19"),
split_by = "Type", relabel = TRUE)
# as well as splitting
ClonalStatPlot(data, group_by = "Subject", groups = c("P17", "P19"),
split_by = "Type", relabel = TRUE)
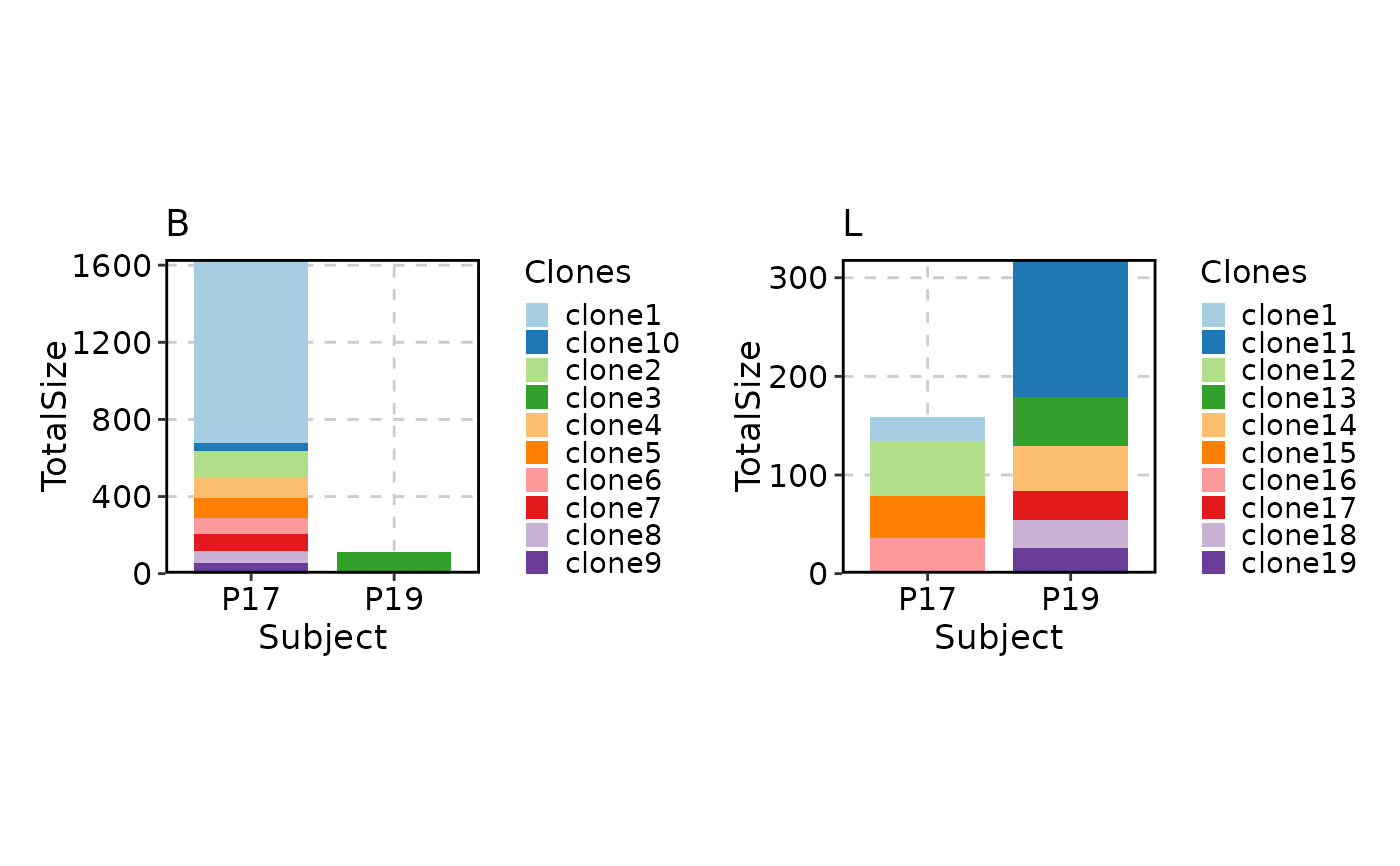 # showing shared clones between P17B and P17L (top 10 clones that are present in both samples)
ClonalStatPlot(data, group_by = "Sample", groups = c("P17B", "P17L"),
clones = "shared(P17B, P17L)", relabel = TRUE, top = 10)
# showing shared clones between P17B and P17L (top 10 clones that are present in both samples)
ClonalStatPlot(data, group_by = "Sample", groups = c("P17B", "P17L"),
clones = "shared(P17B, P17L)", relabel = TRUE, top = 10)
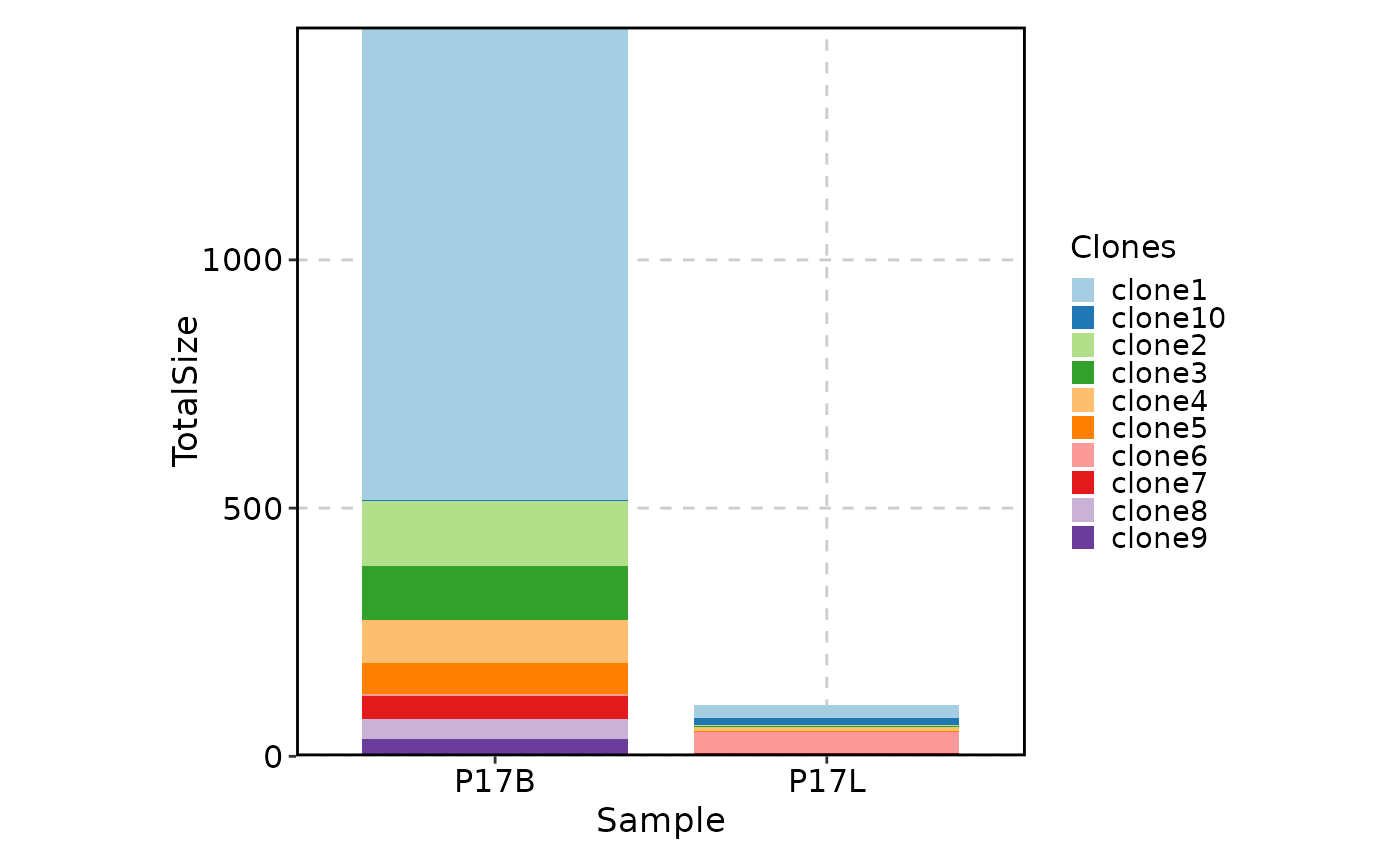 # showing shared clones but with a different order
ClonalStatPlot(data, group_by = "Sample", groups = c("P17B", "P17L"), top = 10,
clones = "shared(P17B, P17L)", relabel = TRUE, orderby = "P17B")
# showing shared clones but with a different order
ClonalStatPlot(data, group_by = "Sample", groups = c("P17B", "P17L"), top = 10,
clones = "shared(P17B, P17L)", relabel = TRUE, orderby = "P17B")
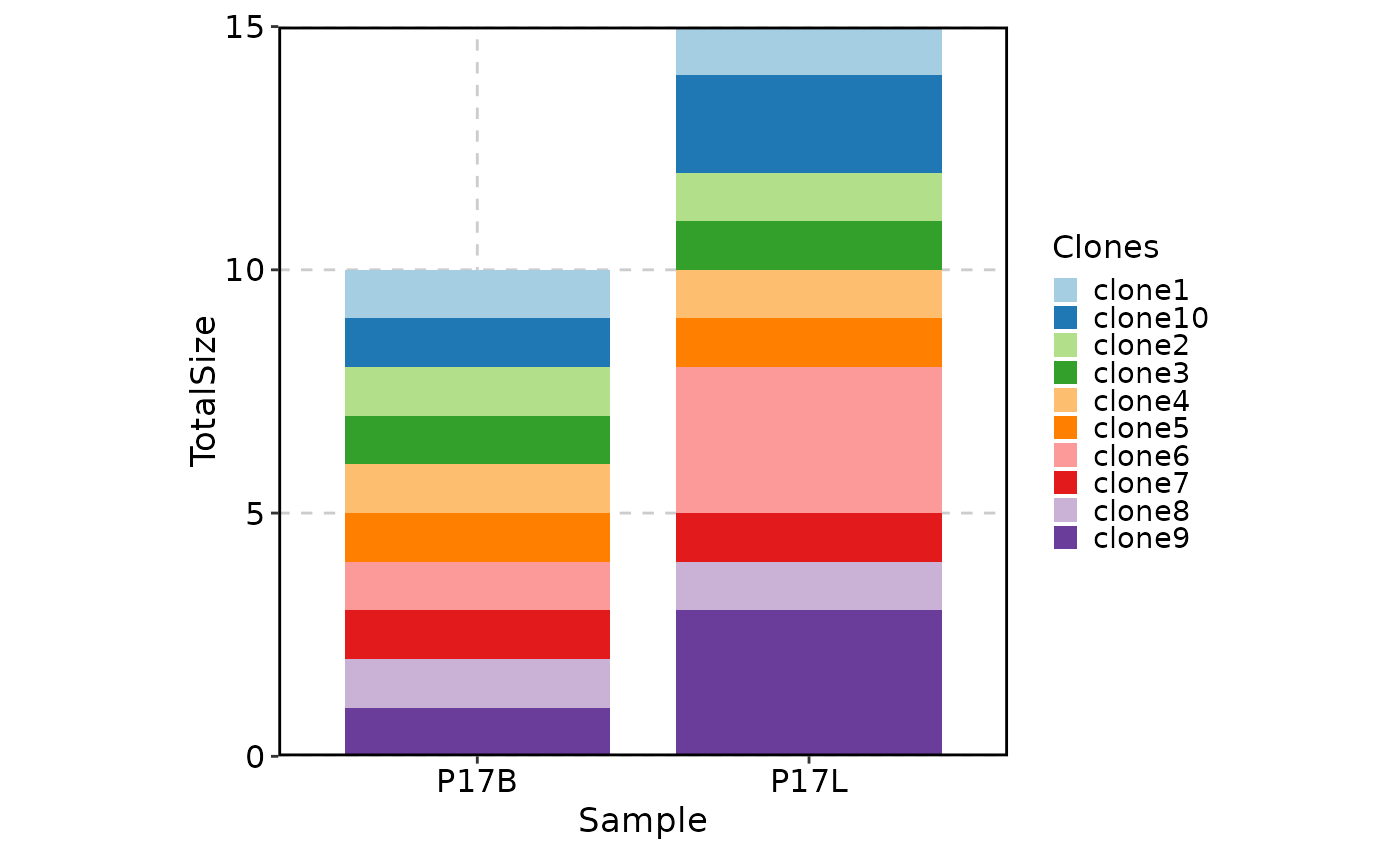 # showing clones larger than 10 in P17L and ordered by the clone size in P17L descendingly
ClonalStatPlot(data, group_by = "Sample", groups = c("P17B", "P17L"),
clones = "sel(P17L > 10)", relabel = TRUE, top = 5, orderby = "P17L")
# showing clones larger than 10 in P17L and ordered by the clone size in P17L descendingly
ClonalStatPlot(data, group_by = "Sample", groups = c("P17B", "P17L"),
clones = "sel(P17L > 10)", relabel = TRUE, top = 5, orderby = "P17L")
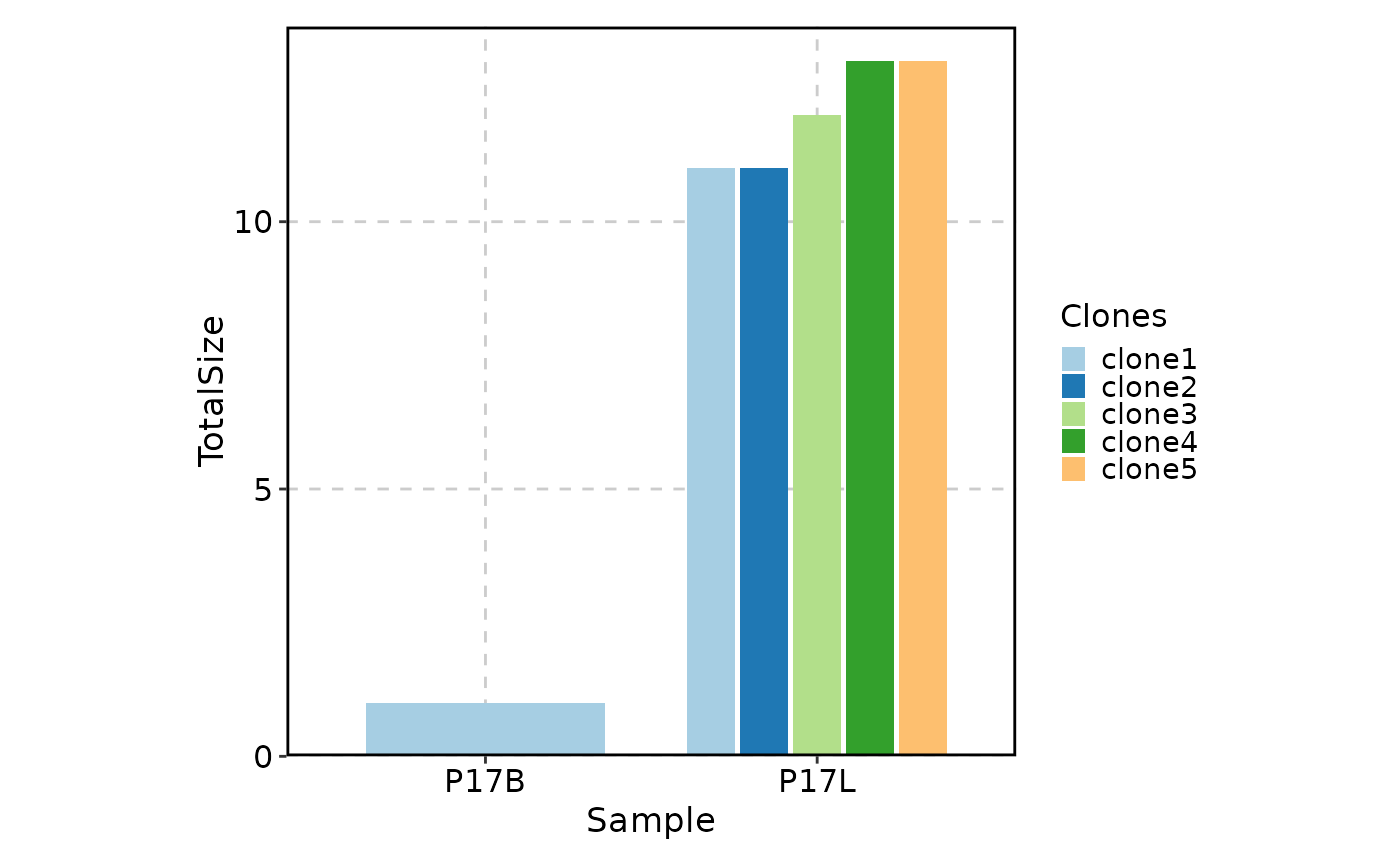 # using trend plot
ClonalStatPlot(data, group_by = "Sample", groups = c("P17B", "P17L"),
clones = sel(P17L > 10 & P17B > 0), relabel = TRUE, orderby = "P17L",
plot_type = "trend")
# using trend plot
ClonalStatPlot(data, group_by = "Sample", groups = c("P17B", "P17L"),
clones = sel(P17L > 10 & P17B > 0), relabel = TRUE, orderby = "P17L",
plot_type = "trend")
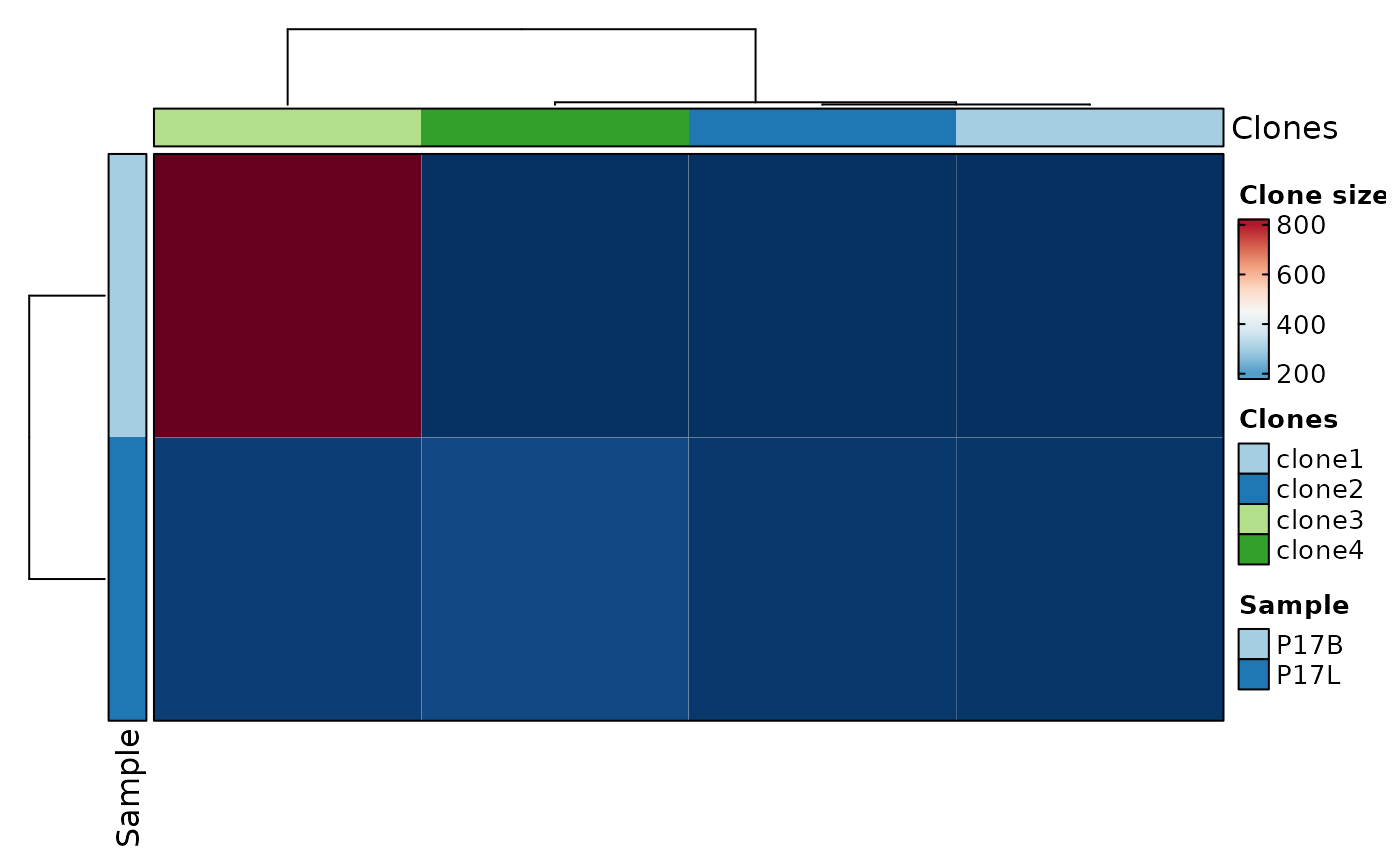
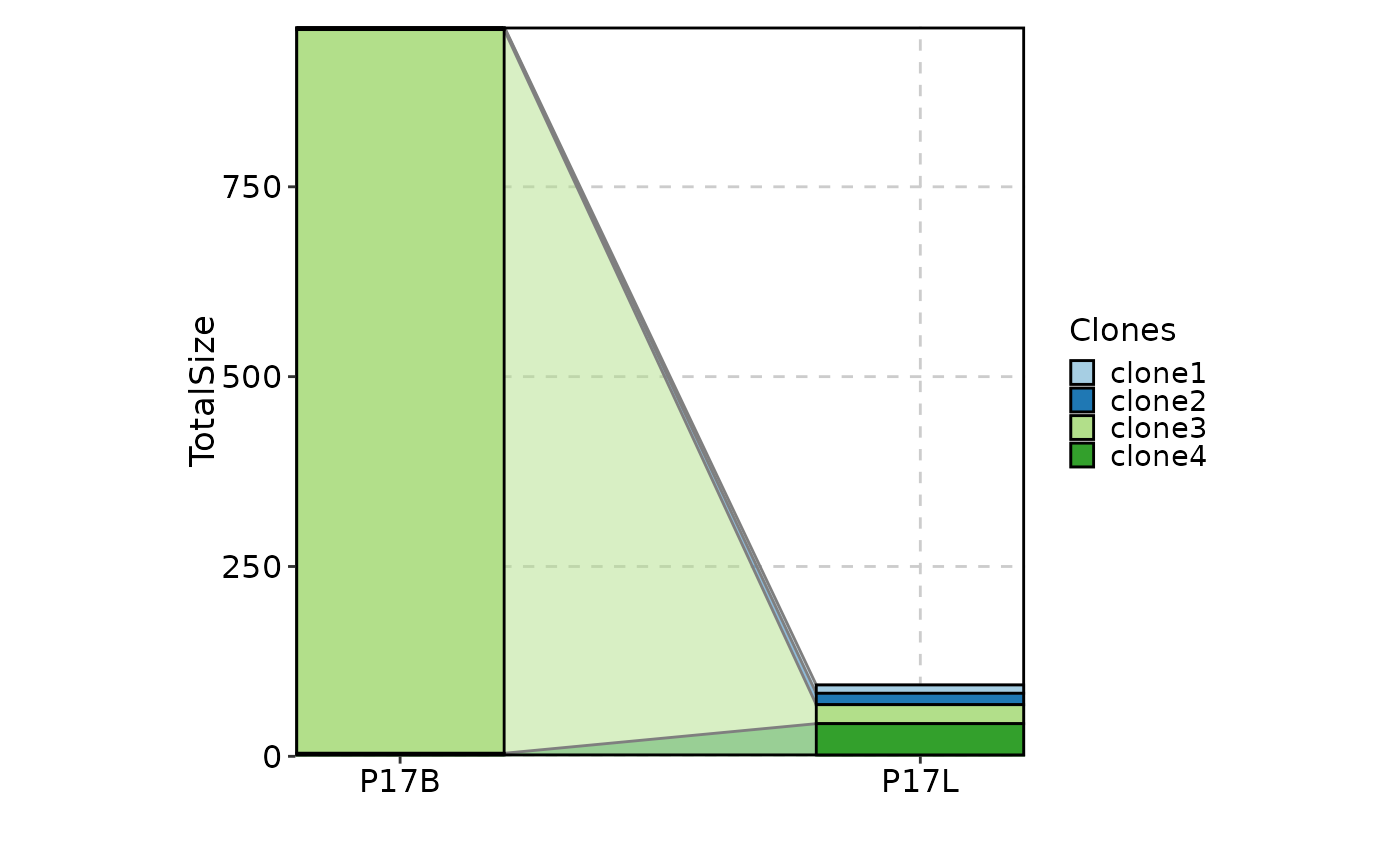 # using heatmap
ClonalStatPlot(data, group_by = "Sample", groups = c("P17B", "P17L"),
clones = sel(P17L > 10 & P17B > 0), relabel = TRUE, orderby = "P17L",
plot_type = "heatmap")
# using heatmap
ClonalStatPlot(data, group_by = "Sample", groups = c("P17B", "P17L"),
clones = sel(P17L > 10 & P17B > 0), relabel = TRUE, orderby = "P17L",
plot_type = "heatmap")
 # using heatmap with subgroups
ClonalStatPlot(data, group_by = "Sample", groups = c("P17B", "P17L"),
clones = list(
ExpandedClonesInP17L = "sel(P17L > 20)",
ExpandedClonesInP17B = "sel(P17B > 20)"
), subgroup_by = "CellType", pie_size = sqrt,
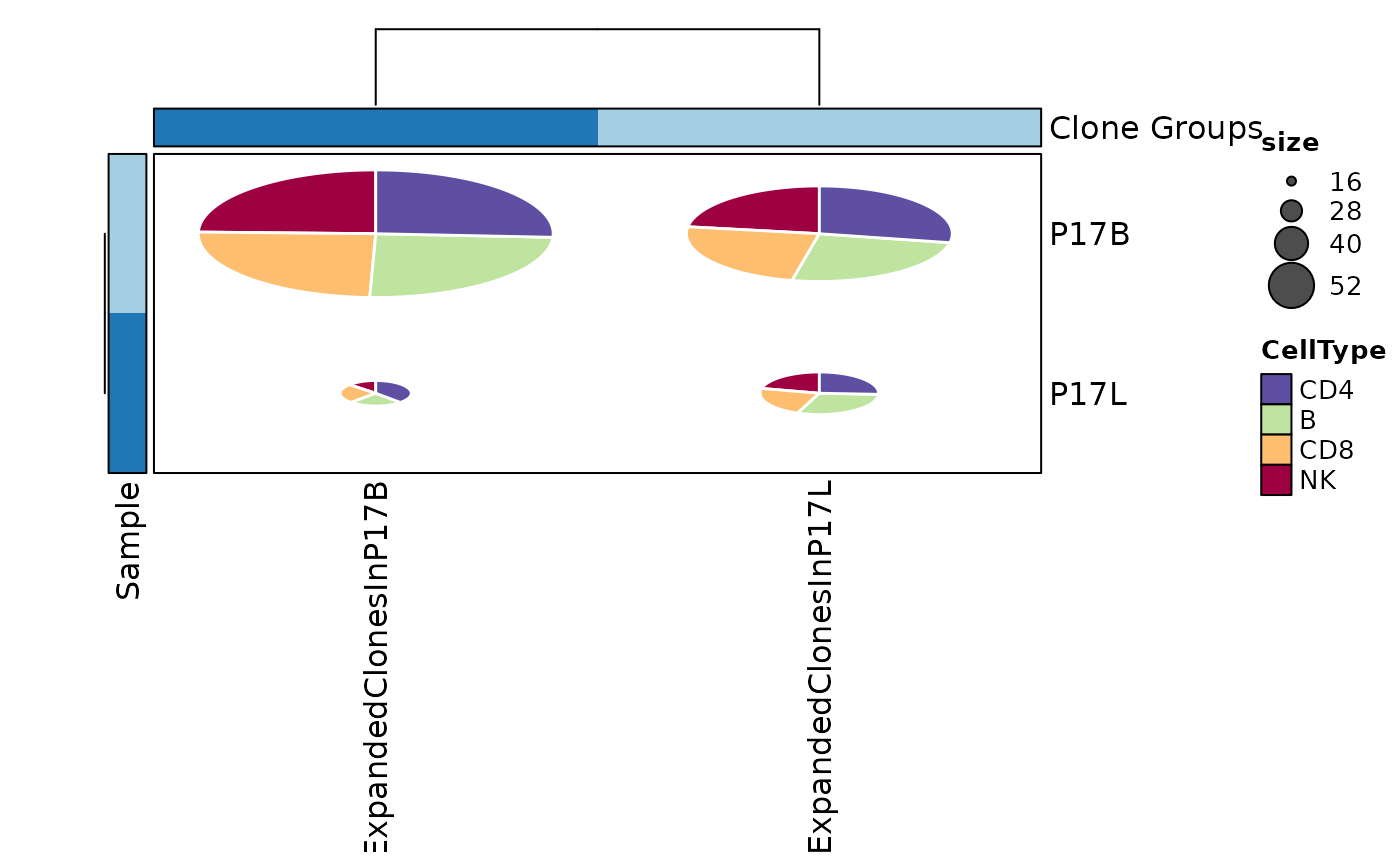
plot_type = "pies", show_row_names = TRUE, show_column_names = TRUE)
# using heatmap with subgroups
ClonalStatPlot(data, group_by = "Sample", groups = c("P17B", "P17L"),
clones = list(
ExpandedClonesInP17L = "sel(P17L > 20)",
ExpandedClonesInP17B = "sel(P17B > 20)"
), subgroup_by = "CellType", pie_size = sqrt,
plot_type = "pies", show_row_names = TRUE, show_column_names = TRUE)
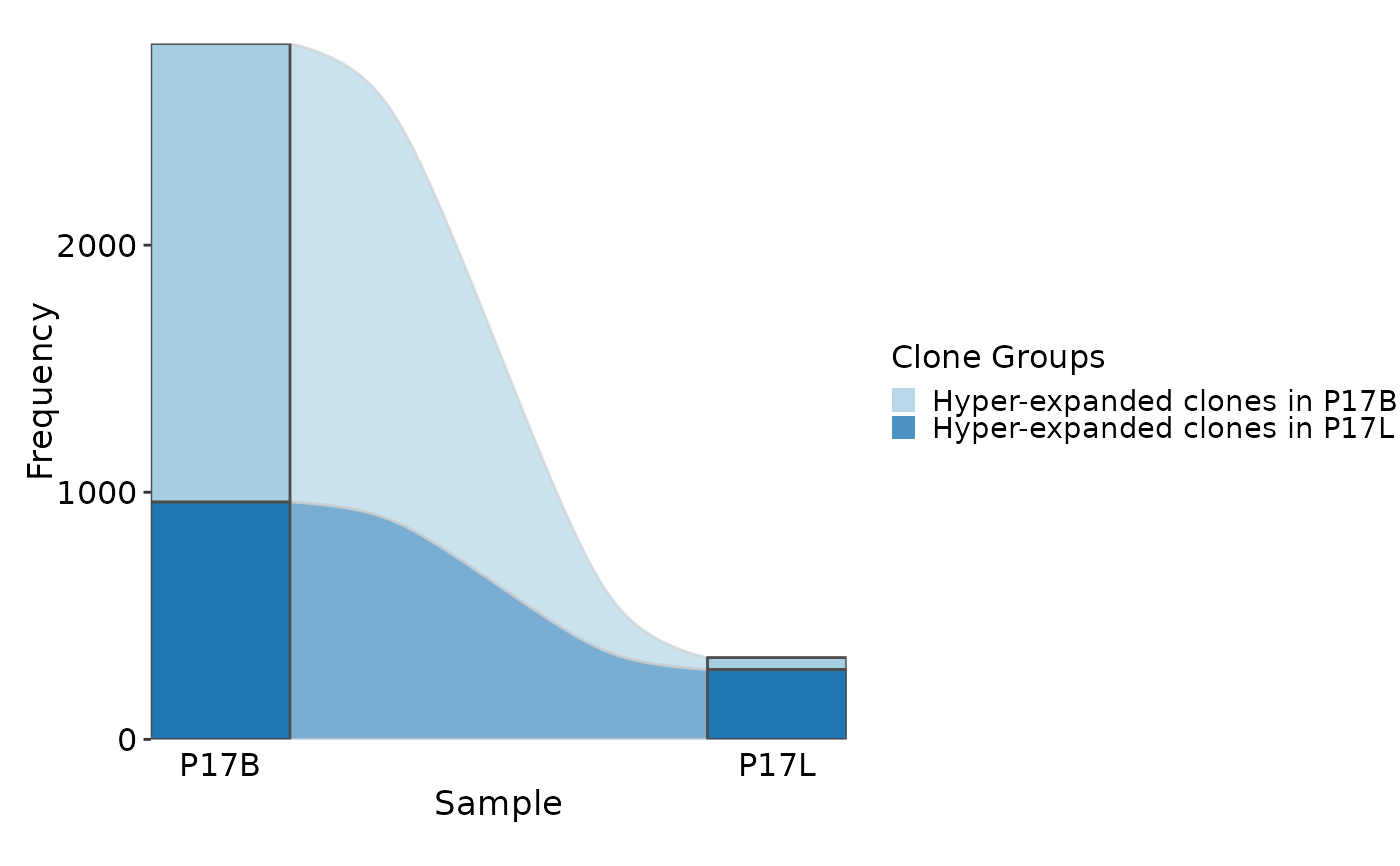
 # using clone groups and showing dynamics using sankey plot
ClonalStatPlot(data, group_by = "Sample", groups = c("P17B", "P17L"),
clones = list(
"Hyper-expanded clones in P17B" = "sel(P17B > 10)",
"Hyper-expanded clones in P17L" = "sel(P17L > 10)"
), plot_type = "sankey")
# using clone groups and showing dynamics using sankey plot
ClonalStatPlot(data, group_by = "Sample", groups = c("P17B", "P17L"),
clones = list(
"Hyper-expanded clones in P17B" = "sel(P17B > 10)",
"Hyper-expanded clones in P17L" = "sel(P17L > 10)"
), plot_type = "sankey")
 # }
# }