A plot visualizing flow/movement/change from one state to another or one time to another.
AlluvialPlot is an alias of SankeyPlot.
Usage
SankeyPlot(
data,
in_form = c("auto", "long", "lodes", "wide", "alluvia", "counts"),
x,
x_sep = "_",
y = NULL,
stratum = NULL,
stratum_sep = "_",
alluvium = NULL,
alluvium_sep = "_",
split_by = NULL,
split_by_sep = "_",
keep_empty = TRUE,
flow = FALSE,
expand = c(0, 0, 0, 0),
nodes_legend = c("auto", "separate", "merge", "none"),
nodes_color = "grey30",
links_fill_by = NULL,
links_fill_by_sep = "_",
links_name = NULL,
links_color = "gray80",
nodes_palette = "Paired",
nodes_palcolor = NULL,
nodes_alpha = 1,
nodes_label = FALSE,
nodes_label_miny = 0,
nodes_width = 0.25,
links_palette = "Paired",
links_palcolor = NULL,
links_alpha = 0.6,
legend.box = "vertical",
x_text_angle = 0,
aspect.ratio = 1,
legend.position = "right",
legend.direction = "vertical",
flip = FALSE,
theme = "theme_this",
theme_args = list(),
title = NULL,
subtitle = NULL,
xlab = NULL,
ylab = NULL,
facet_by = NULL,
facet_scales = "fixed",
facet_ncol = NULL,
facet_nrow = NULL,
facet_byrow = TRUE,
seed = 8525,
combine = TRUE,
nrow = NULL,
ncol = NULL,
byrow = TRUE,
axes = NULL,
axis_titles = axes,
guides = NULL,
design = NULL,
...
)
AlluvialPlot(
data,
in_form = c("auto", "long", "lodes", "wide", "alluvia", "counts"),
x,
x_sep = "_",
y = NULL,
stratum = NULL,
stratum_sep = "_",
alluvium = NULL,
alluvium_sep = "_",
split_by = NULL,
split_by_sep = "_",
keep_empty = TRUE,
flow = FALSE,
expand = c(0, 0, 0, 0),
nodes_legend = c("auto", "separate", "merge", "none"),
nodes_color = "grey30",
links_fill_by = NULL,
links_fill_by_sep = "_",
links_name = NULL,
links_color = "gray80",
nodes_palette = "Paired",
nodes_palcolor = NULL,
nodes_alpha = 1,
nodes_label = FALSE,
nodes_label_miny = 0,
nodes_width = 0.25,
links_palette = "Paired",
links_palcolor = NULL,
links_alpha = 0.6,
legend.box = "vertical",
x_text_angle = 0,
aspect.ratio = 1,
legend.position = "right",
legend.direction = "vertical",
flip = FALSE,
theme = "theme_this",
theme_args = list(),
title = NULL,
subtitle = NULL,
xlab = NULL,
ylab = NULL,
facet_by = NULL,
facet_scales = "fixed",
facet_ncol = NULL,
facet_nrow = NULL,
facet_byrow = TRUE,
seed = 8525,
combine = TRUE,
nrow = NULL,
ncol = NULL,
byrow = TRUE,
axes = NULL,
axis_titles = axes,
guides = NULL,
design = NULL,
...
)Arguments
- data
A data frame in following possible formats:
"long" or "lodes": A long format with columns for
x,stratum,alluvium, andy.x(required, single columns or concatenated byx_sep) is the column name to plot on the x-axis,stratum(defaults tolinks_fill_by) is the column name to group the nodes for eachx,alluvium(required) is the column name to define the links, andyis the frequency of eachx,stratum, andalluvium."wide" or "alluvia": A wide format with columns for
x.x(required, multiple columns,x_sepwon't be used) are the columns to plot on the x-axis,stratumandalluviumwill be ignored. See ggalluvial::to_lodes_form for more details."counts": A format with counts being provides under each
x.x(required, multiple columns,x_sepwon't be used) are the columns to plot on the x-axis. When the first element ofxis ".", values oflinks_fill_by(required) will be added to the plot as the first column of nodes. It is useful to show how the links are flowed from the source to the targets."auto" (default): Automatically determine the format based on the columns provided. When the length of
xis greater than 1 and allxcolumns are numeric, "counts" format will be used. When the length ofxis greater than 1 and ggalluvial::is_alluvia_form returns TRUE, "alluvia" format will be used. Otherwise, "lodes" format will be tried.
- in_form
A character string to specify the format of the data. Possible values are "auto", "long", "lodes", "wide", "alluvia", and "counts".
- x
A character string specifying the column name of the data frame to plot for the x-axis.
- x_sep
A character string to concatenate the columns in
x, if multiple columns are provided.- y
A character string specifying the column name of the data frame to plot for the y-axis.
- stratum
A character string of the column name to group the nodes for each
x. Seedatafor more details.- stratum_sep
A character string to concatenate the columns in
stratum, if multiple columns are provided.- alluvium
A character string of the column name to define the links. See
datafor more details.- alluvium_sep
A character string to concatenate the columns in
alluvium, if multiple columns are provided.- split_by
The column(s) to split data by and plot separately.
- split_by_sep
The separator for multiple split_by columns. See
split_by- keep_empty
One of FALSE, TRUE and "level". It can also take a named list to specify different behavior for different columns. Without a named list, the behavior applies to the categorical/character columns used on the plot, for example, the
x,group_by,fill_by, etc.FALSE(default): Drop empty factor levels from the data before plotting.TRUE: Keep empty factor levels and show them as a separate category in the plot."level": Keep empty factor levels, but do not show them in the plot. But they will be assigned colors from the palette to maintain consistency across multiple plots. Alias:levels
- flow
A logical value to use ggalluvial::geom_flow instead of ggalluvial::geom_alluvium.
- expand
The values to expand the x and y axes. It is like CSS padding. When a single value is provided, it is used for both axes on both sides. When two values are provided, the first value is used for the top/bottom side and the second value is used for the left/right side. When three values are provided, the first value is used for the top side, the second value is used for the left/right side, and the third value is used for the bottom side. When four values are provided, the values are used for the top, right, bottom, and left sides, respectively. You can also use a named vector to specify the values for each side. When the axis is discrete, the values will be applied as 'add' to the 'expansion' function. When the axis is continuous, the values will be applied as 'mult' to the 'expansion' function. See also https://ggplot2.tidyverse.org/reference/expansion.html
- nodes_legend
Controls how the legend of nodes will be shown. Possible values are:
"merge": Merge the legends of nodes. That is only one legend will be shown for all nodes.
"separate": Show the legends of nodes separately. That is, nodes on each
xwill have their own legend."none": Do not show the legend of nodes.
"auto": Automatically determine how to show the legend. When
nodes_labelis TRUE, "none" will apply. Whennodes_labelis FALSE, and if stratum is the same as links_fill_by, "none" will apply. If there is any overlapping values between the nodes on differentx, "merge" will apply. Otherwise, "separate" will apply.
- nodes_color
A character string to color the nodes. Use a special value ".fill" to use the same color as the fill.
- links_fill_by
A character string of the column name to fill the links.
- links_fill_by_sep
A character string to concatenate the columns in
links_fill_by, if multiple columns are provided.- links_name
A character string to name the legend of links.
- links_color
A character string to color the borders of links. Use a special value ".fill" to use the same color as the fill.
- nodes_palette
A character string to specify the palette of nodes fill.
- nodes_palcolor
A character vector to specify the colors of nodes fill.
- nodes_alpha
A numeric value to specify the transparency of nodes fill.
- nodes_label
A logical value to show the labels on the nodes.
- nodes_label_miny
A numeric value to specify the minimum y (frequency) to show the labels.
- nodes_width
A numeric value to specify the width of nodes.
- links_palette
A character string to specify the palette of links fill.
- links_palcolor
A character vector to specify the colors of links fill.
- links_alpha
A numeric value to specify the transparency of links fill.
- legend.box
A character string to specify the box of the legend, either "vertical" or "horizontal".
- x_text_angle
A numeric value specifying the angle of the x-axis text.
- aspect.ratio
A numeric value specifying the aspect ratio of the plot.
- legend.position
A character string specifying the position of the legend. if
waiver(), for single groups, the legend will be "none", otherwise "right".- legend.direction
A character string specifying the direction of the legend.
- flip
A logical value to flip the plot.
- theme
A character string or a theme class (i.e. ggplot2::theme_classic) specifying the theme to use. Default is "theme_this".
- theme_args
A list of arguments to pass to the theme function.
- title
A character string specifying the title of the plot. A function can be used to generate the title based on the default title. This is useful when split_by is used and the title needs to be dynamic.
- subtitle
A character string specifying the subtitle of the plot.
- xlab
A character string specifying the x-axis label.
- ylab
A character string specifying the y-axis label.
- facet_by
A character string specifying the column name of the data frame to facet the plot. Otherwise, the data will be split by
split_byand generate multiple plots and combine them into one usingpatchwork::wrap_plots- facet_scales
Whether to scale the axes of facets. Default is "fixed" Other options are "free", "free_x", "free_y". See
ggplot2::facet_wrap- facet_ncol
A numeric value specifying the number of columns in the facet. When facet_by is a single column and facet_wrap is used.
- facet_nrow
A numeric value specifying the number of rows in the facet. When facet_by is a single column and facet_wrap is used.
- facet_byrow
A logical value indicating whether to fill the plots by row. Default is TRUE.
- seed
The random seed to use. Default is 8525.
- combine
Whether to combine the plots into one when facet is FALSE. Default is TRUE.
- nrow
A numeric value specifying the number of rows in the facet.
- ncol
A numeric value specifying the number of columns in the facet.
- byrow
A logical value indicating whether to fill the plots by row.
- axes
A string specifying how axes should be treated. Passed to
patchwork::wrap_plots(). Only relevant whensplit_byis used andcombineis TRUE. Options are:'keep' will retain all axes in individual plots.
'collect' will remove duplicated axes when placed in the same run of rows or columns of the layout.
'collect_x' and 'collect_y' will remove duplicated x-axes in the columns or duplicated y-axes in the rows respectively.
- axis_titles
A string specifying how axis titltes should be treated. Passed to
patchwork::wrap_plots(). Only relevant whensplit_byis used andcombineis TRUE. Options are:'keep' will retain all axis titles in individual plots.
'collect' will remove duplicated titles in one direction and merge titles in the opposite direction.
'collect_x' and 'collect_y' control this for x-axis titles and y-axis titles respectively.
- guides
A string specifying how guides should be treated in the layout. Passed to
patchwork::wrap_plots(). Only relevant whensplit_byis used andcombineis TRUE. Options are:'collect' will collect guides below to the given nesting level, removing duplicates.
'keep' will stop collection at this level and let guides be placed alongside their plot.
'auto' will allow guides to be collected if a upper level tries, but place them alongside the plot if not.
- design
Specification of the location of areas in the layout, passed to
patchwork::wrap_plots(). Only relevant whensplit_byis used andcombineis TRUE. When specified,nrow,ncol, andbyroware ignored. Seepatchwork::wrap_plots()for more details.- ...
Additional arguments.
Examples
# \donttest{
# Reproduce the examples in ggalluvial
set.seed(8525)
data(UCBAdmissions, package = "datasets")
UCBAdmissions <- as.data.frame(UCBAdmissions)
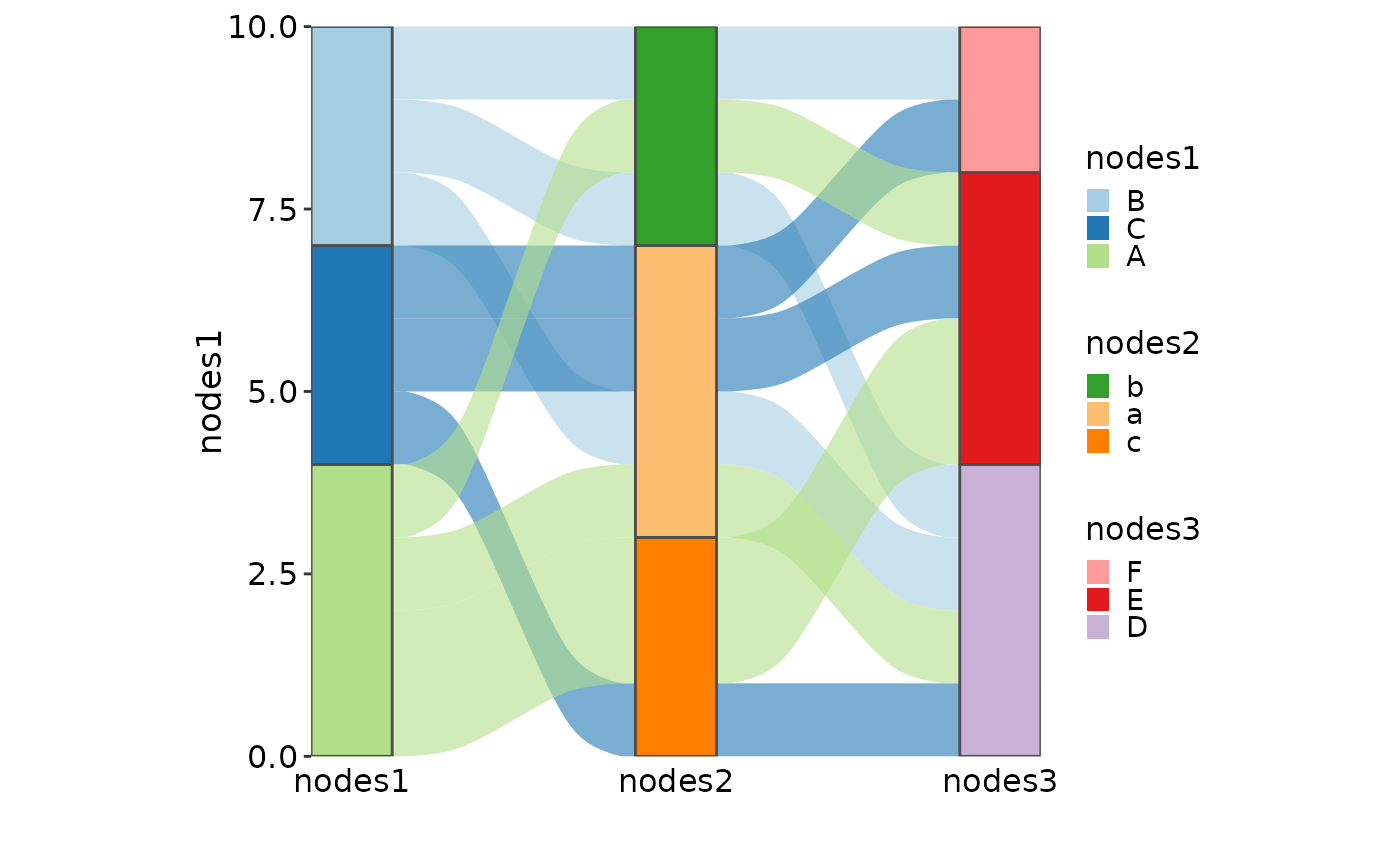
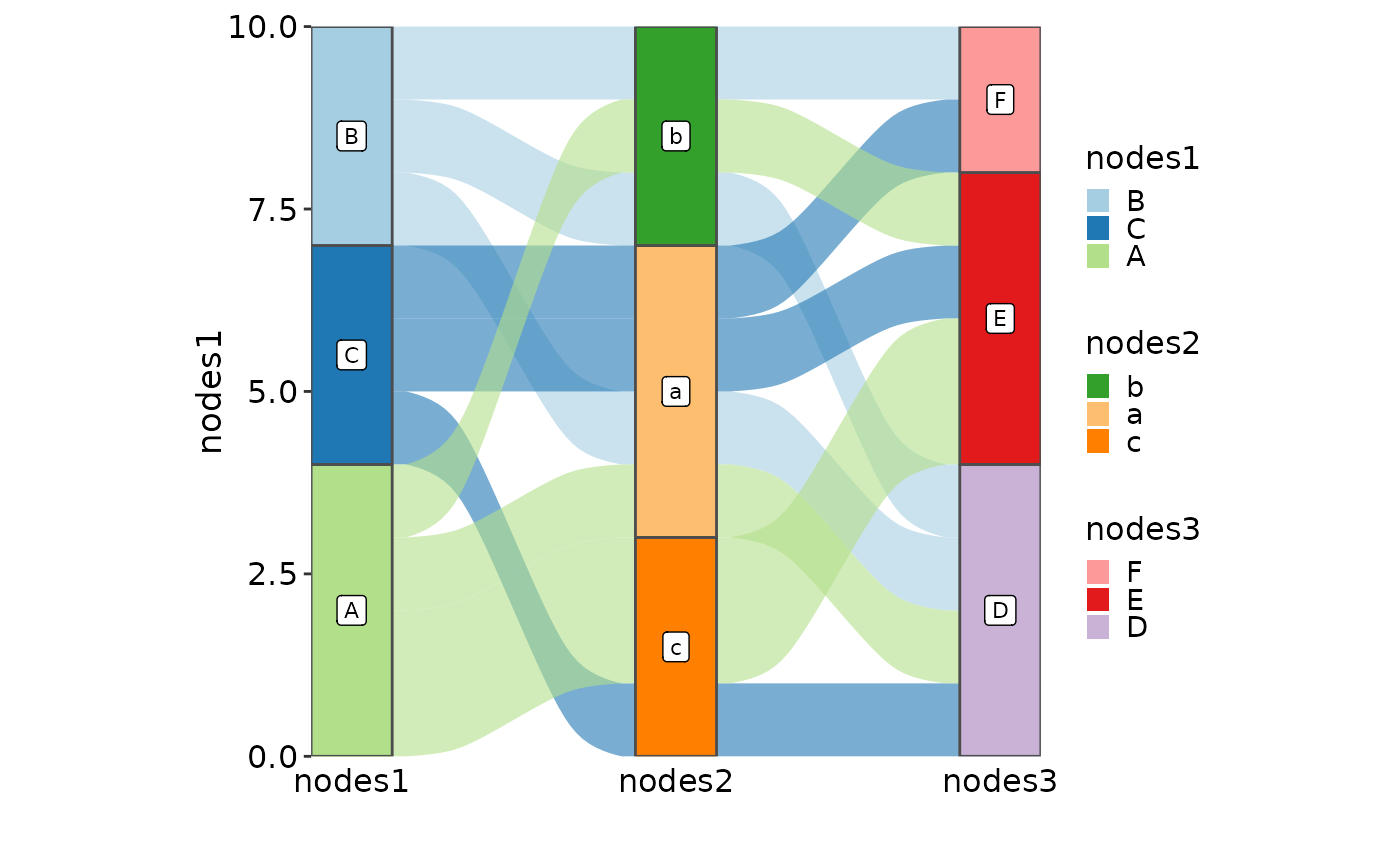
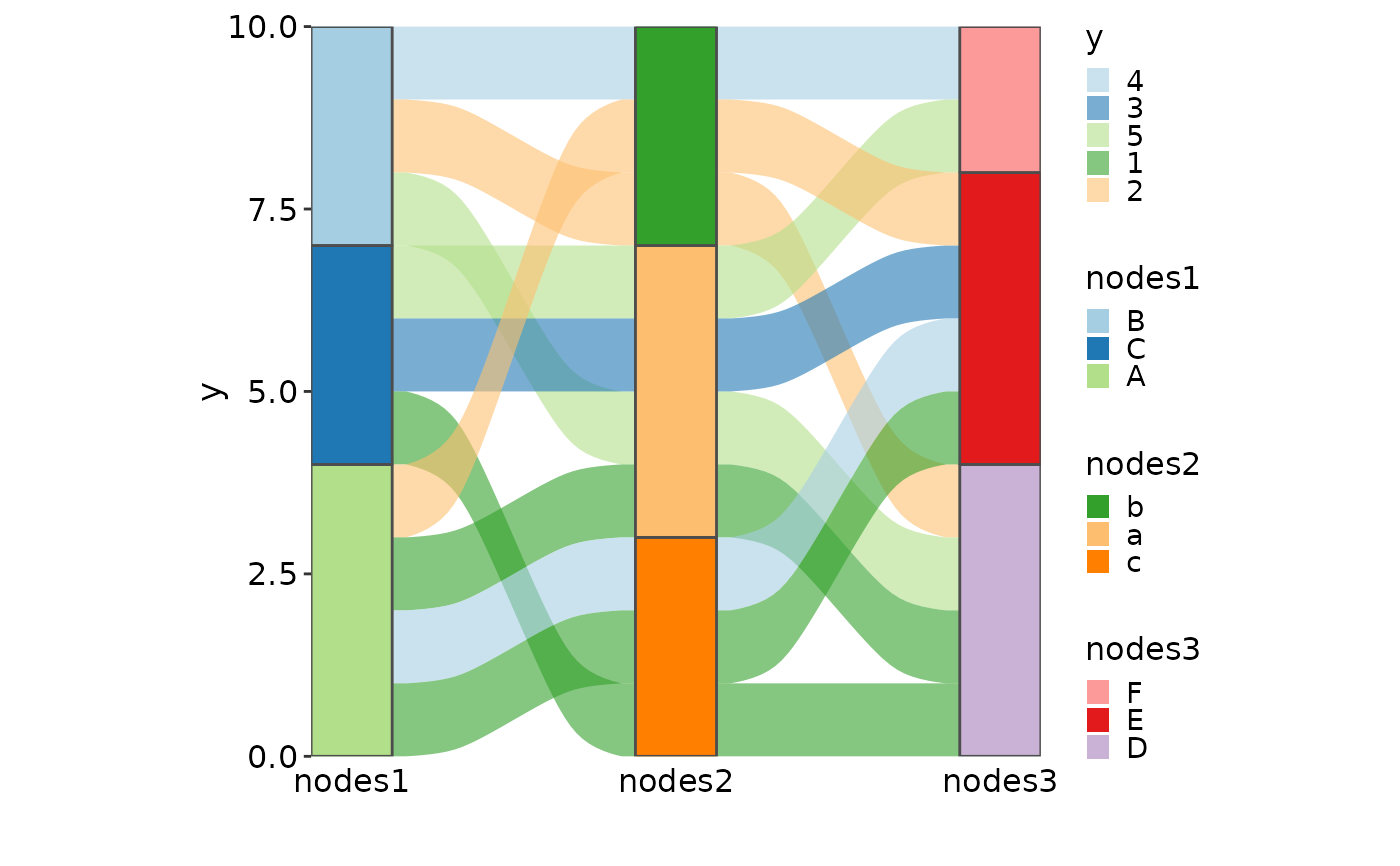
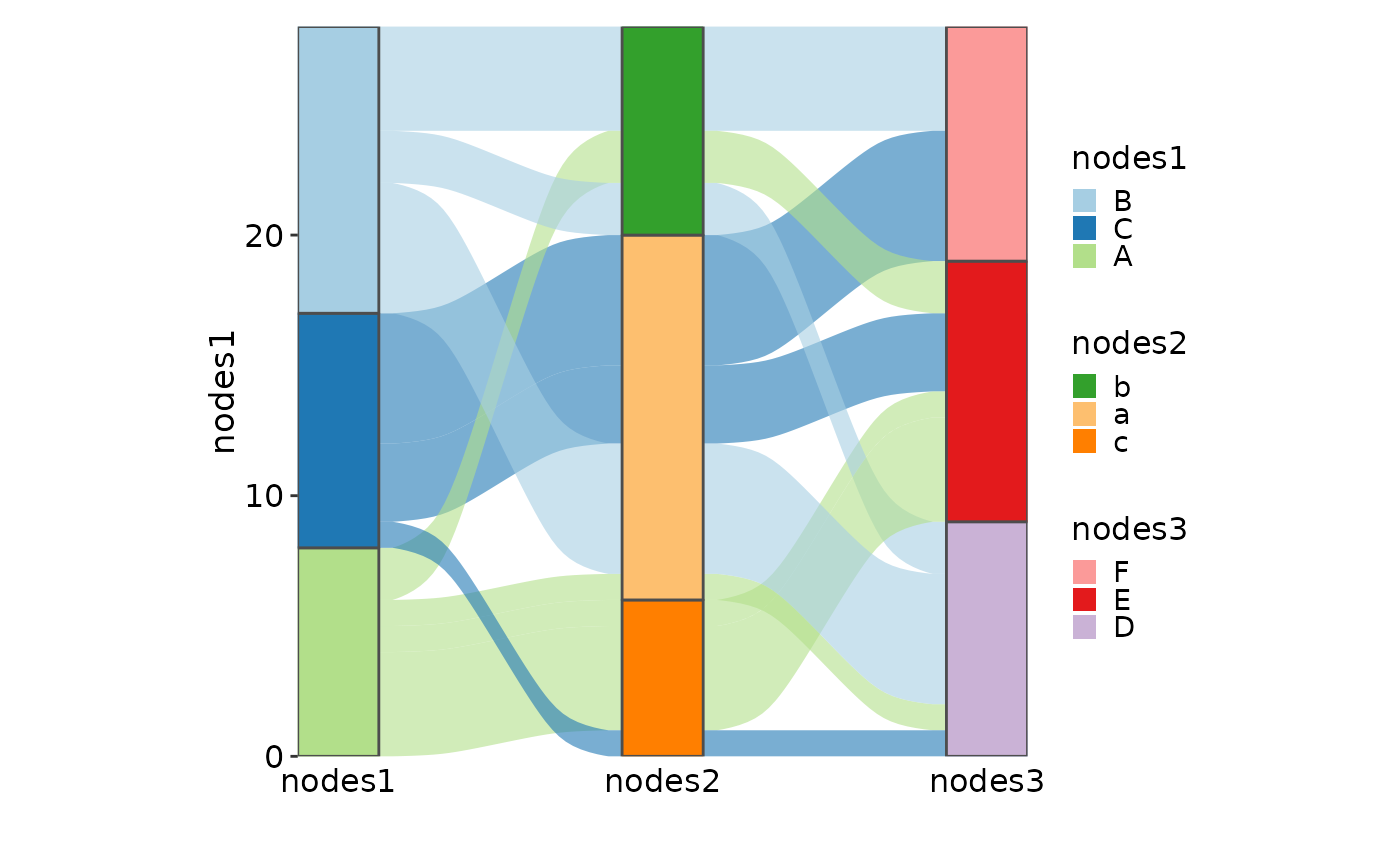
SankeyPlot(as.data.frame(UCBAdmissions), x = c("Gender", "Dept"),
y = "Freq", nodes_width = 1/12, links_fill_by = "Admit", nodes_label = TRUE,
nodes_palette = "simspec", links_palette = "Set1", links_alpha = 0.5,
nodes_palcolor = "black", links_color = "transparent")
 data(HairEyeColor, package = "datasets")
SankeyPlot(as.data.frame(HairEyeColor), x = c("Hair", "Eye", "Sex"),
y = "Freq", links_fill_by = "Eye", nodes_width = 1/8, nodes_alpha = 0.4,
flip = TRUE, reverse = FALSE, knot.pos = 0, links_color = "transparent",
ylab = "Freq", links_alpha = 0.5, links_name = "Eye (links)", links_palcolor = c(
Brown = "#70493D", Hazel = "#E2AC76", Green = "#3F752B", Blue = "#81B0E4"))
data(HairEyeColor, package = "datasets")
SankeyPlot(as.data.frame(HairEyeColor), x = c("Hair", "Eye", "Sex"),
y = "Freq", links_fill_by = "Eye", nodes_width = 1/8, nodes_alpha = 0.4,
flip = TRUE, reverse = FALSE, knot.pos = 0, links_color = "transparent",
ylab = "Freq", links_alpha = 0.5, links_name = "Eye (links)", links_palcolor = c(
Brown = "#70493D", Hazel = "#E2AC76", Green = "#3F752B", Blue = "#81B0E4"))
 data(Refugees, package = "alluvial")
country_regions <- c(
Afghanistan = "Middle East",
Burundi = "Central Africa",
`Congo DRC` = "Central Africa",
Iraq = "Middle East",
Myanmar = "Southeast Asia",
Palestine = "Middle East",
Somalia = "Horn of Africa",
Sudan = "Central Africa",
Syria = "Middle East",
Vietnam = "Southeast Asia"
)
Refugees$region <- country_regions[Refugees$country]
SankeyPlot(Refugees, x = "year", y = "refugees", alluvium = "country",
links_fill_by = "country", links_color = ".fill", links_alpha = 0.75,
links_palette = "Set3", facet_by = "region", x_text_angle = -45, nodes_legend = "none",
theme_args = list(strip.background = ggplot2::element_rect(fill="grey80")),
decreasing = FALSE, nodes_width = 0, nodes_color = "transparent", ylab = "refugees",
title = "Refugee volume by country and region of origin")
data(Refugees, package = "alluvial")
country_regions <- c(
Afghanistan = "Middle East",
Burundi = "Central Africa",
`Congo DRC` = "Central Africa",
Iraq = "Middle East",
Myanmar = "Southeast Asia",
Palestine = "Middle East",
Somalia = "Horn of Africa",
Sudan = "Central Africa",
Syria = "Middle East",
Vietnam = "Southeast Asia"
)
Refugees$region <- country_regions[Refugees$country]
SankeyPlot(Refugees, x = "year", y = "refugees", alluvium = "country",
links_fill_by = "country", links_color = ".fill", links_alpha = 0.75,
links_palette = "Set3", facet_by = "region", x_text_angle = -45, nodes_legend = "none",
theme_args = list(strip.background = ggplot2::element_rect(fill="grey80")),
decreasing = FALSE, nodes_width = 0, nodes_color = "transparent", ylab = "refugees",
title = "Refugee volume by country and region of origin")
 data(majors, package = "ggalluvial")
majors$curriculum <- as.factor(majors$curriculum)
SankeyPlot(majors, x = "semester", stratum = "curriculum", alluvium = "student",
links_fill_by = "curriculum", flow = TRUE, stat = "alluvium", nodes_palette = "Set2",
links_palette = "Set2")
data(majors, package = "ggalluvial")
majors$curriculum <- as.factor(majors$curriculum)
SankeyPlot(majors, x = "semester", stratum = "curriculum", alluvium = "student",
links_fill_by = "curriculum", flow = TRUE, stat = "alluvium", nodes_palette = "Set2",
links_palette = "Set2")
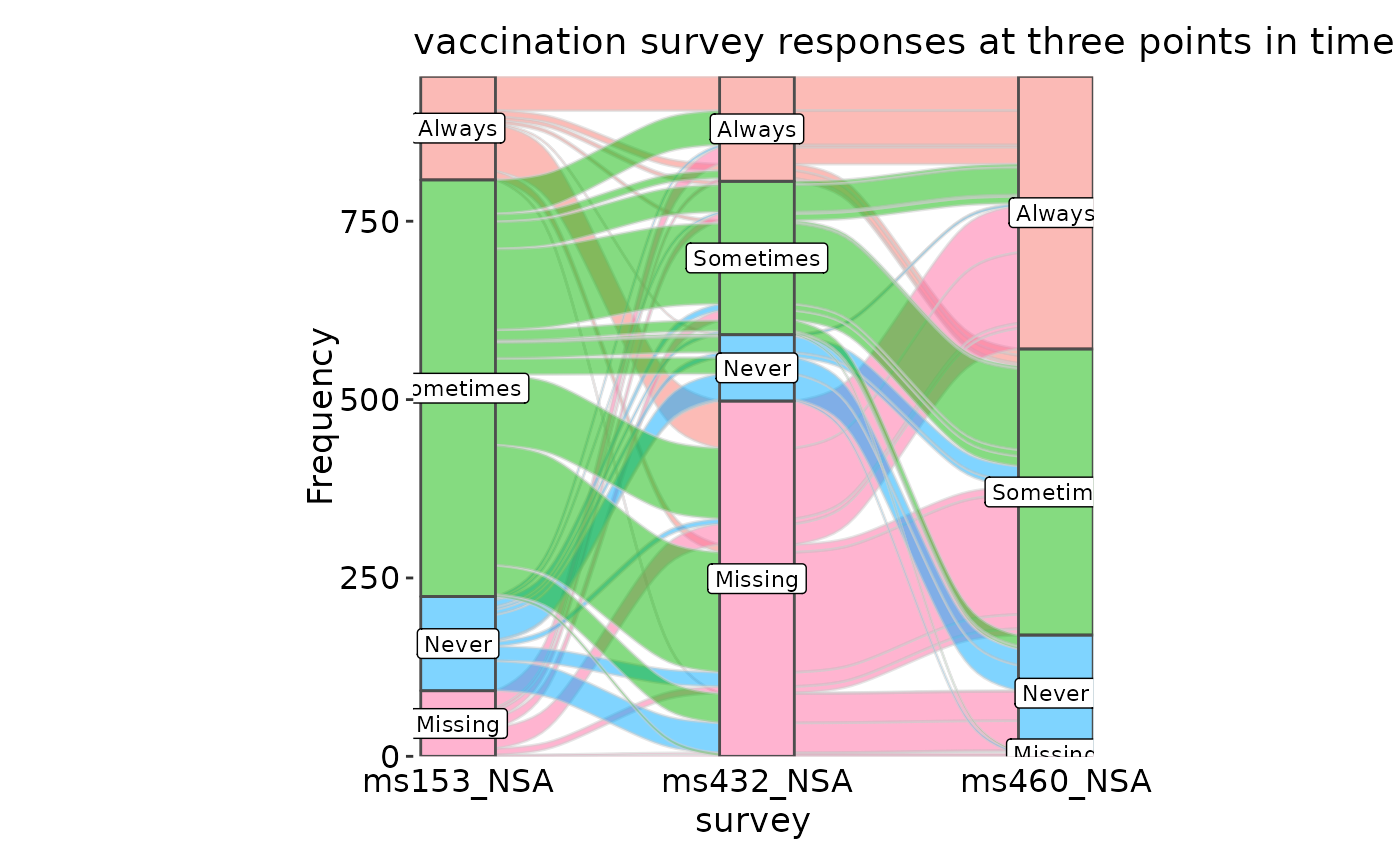
 data(vaccinations, package = "ggalluvial")
vaccinations <- transform(vaccinations,
response = factor(response, rev(levels(response))))
SankeyPlot(vaccinations, x = "survey", stratum = "response", alluvium = "subject",
y = "freq", links_fill_by = "response", nodes_label = TRUE, nodes_alpha = 0.5,
nodes_palette = "seurat", links_palette = "seurat", links_alpha = 0.5,
legend.position = "none", flow = TRUE, expand = c(0, 0, 0, .15), stat = "alluvium",
title = "vaccination survey responses at three points in time")
data(vaccinations, package = "ggalluvial")
vaccinations <- transform(vaccinations,
response = factor(response, rev(levels(response))))
SankeyPlot(vaccinations, x = "survey", stratum = "response", alluvium = "subject",
y = "freq", links_fill_by = "response", nodes_label = TRUE, nodes_alpha = 0.5,
nodes_palette = "seurat", links_palette = "seurat", links_alpha = 0.5,
legend.position = "none", flow = TRUE, expand = c(0, 0, 0, .15), stat = "alluvium",
title = "vaccination survey responses at three points in time")
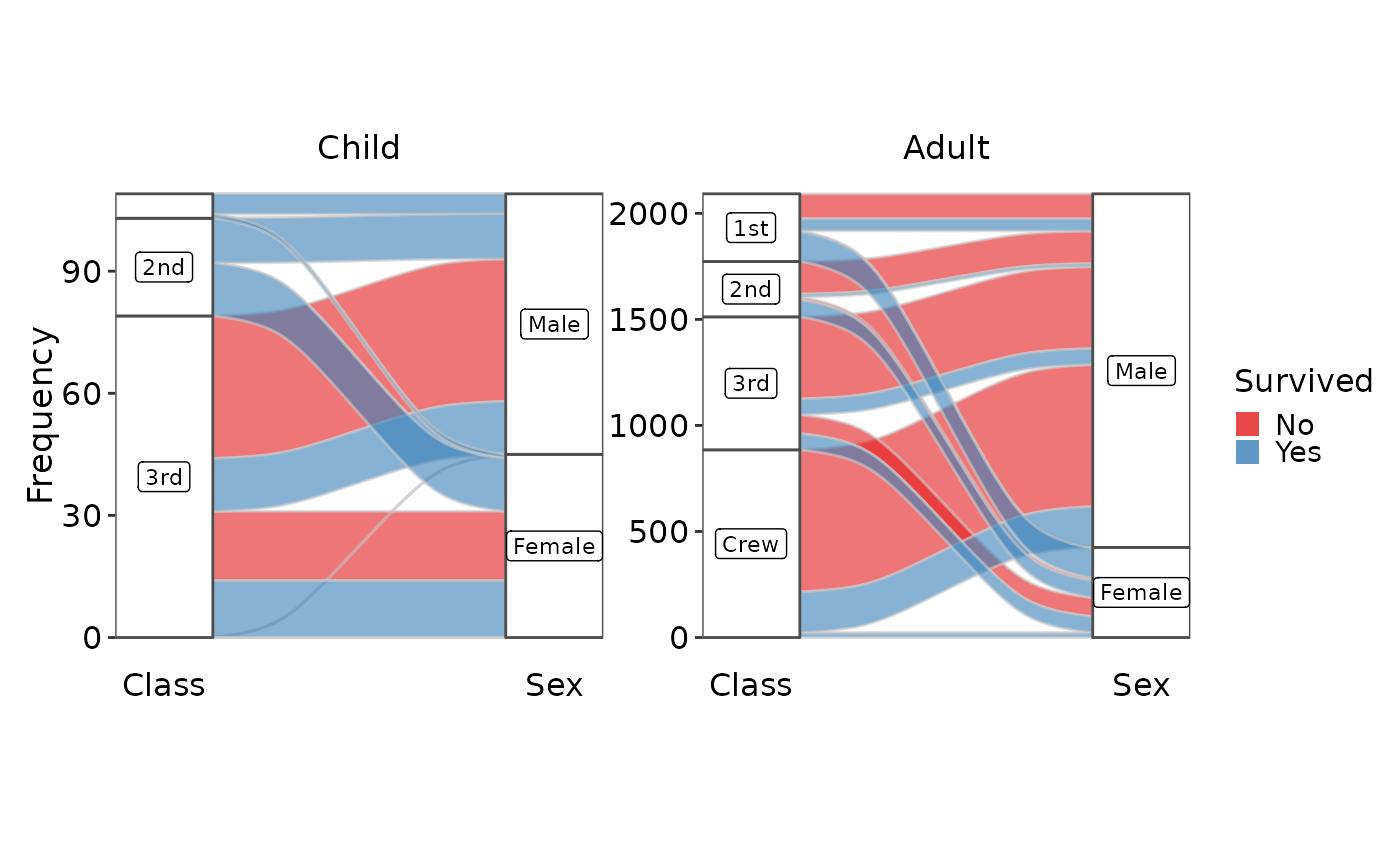
 data(Titanic, package = "datasets")
SankeyPlot(as.data.frame(Titanic), x = c("Class", "Sex"), y = "Freq",
links_fill_by = "Survived", flow = TRUE, facet_by = "Age", facet_scales = "free_y",
nodes_label = TRUE, expand = c(0.05, 0), xlab = "", links_palette = "Set1",
nodes_palcolor = "white", nodes_label_miny = 10)
data(Titanic, package = "datasets")
SankeyPlot(as.data.frame(Titanic), x = c("Class", "Sex"), y = "Freq",
links_fill_by = "Survived", flow = TRUE, facet_by = "Age", facet_scales = "free_y",
nodes_label = TRUE, expand = c(0.05, 0), xlab = "", links_palette = "Set1",
nodes_palcolor = "white", nodes_label_miny = 10)
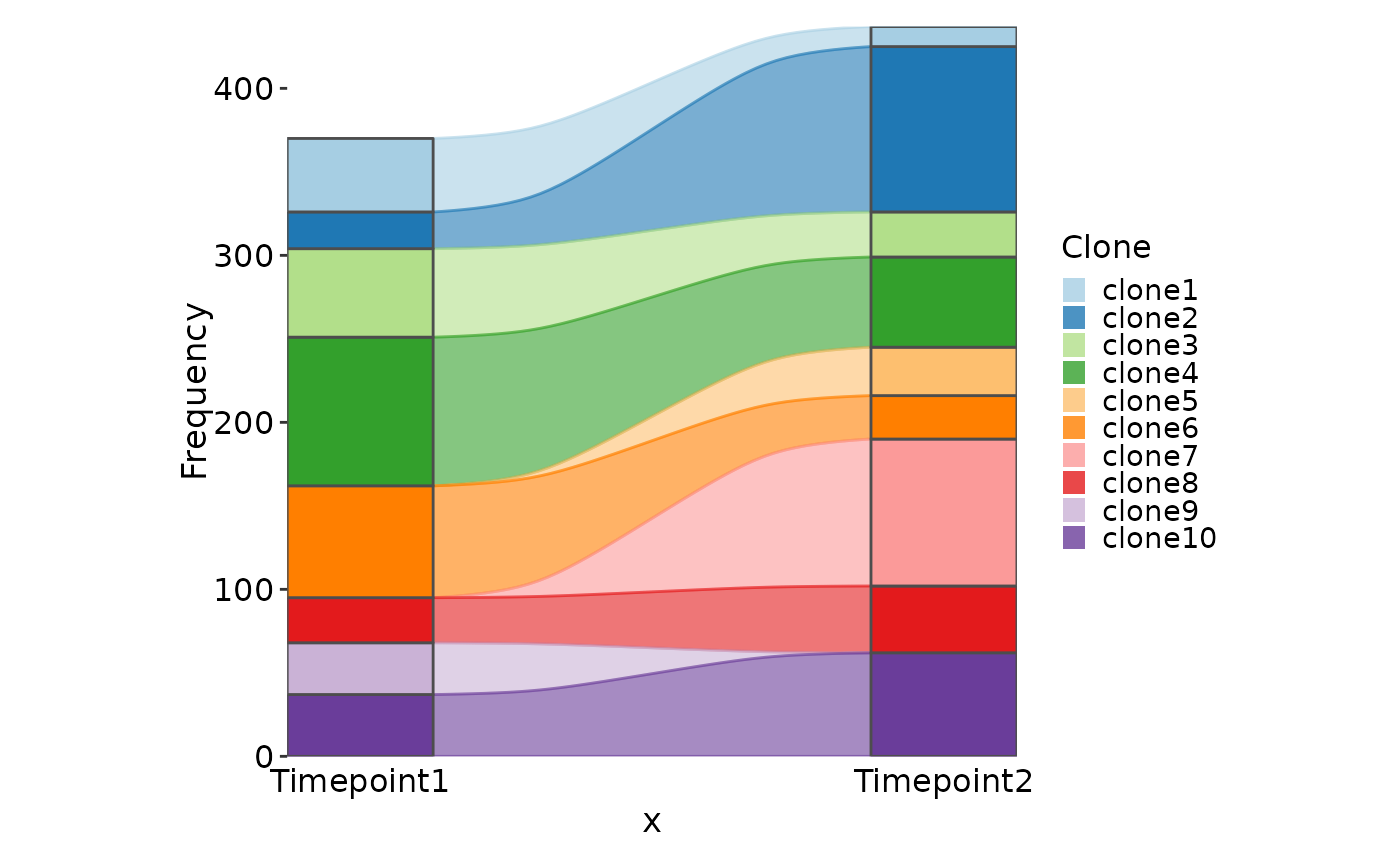
 # Simulated examples
df <- data.frame(
Clone = paste0("clone", 1:10),
Timepoint1 = sample(c(rep(0, 30), 1:100), 10),
Timepoint2 = sample(c(rep(0, 30), 1:100), 10)
)
SankeyPlot(df, x = c("Timepoint1", "Timepoint2"), alluvium = "Clone",
links_color = ".fill")
# Simulated examples
df <- data.frame(
Clone = paste0("clone", 1:10),
Timepoint1 = sample(c(rep(0, 30), 1:100), 10),
Timepoint2 = sample(c(rep(0, 30), 1:100), 10)
)
SankeyPlot(df, x = c("Timepoint1", "Timepoint2"), alluvium = "Clone",
links_color = ".fill")
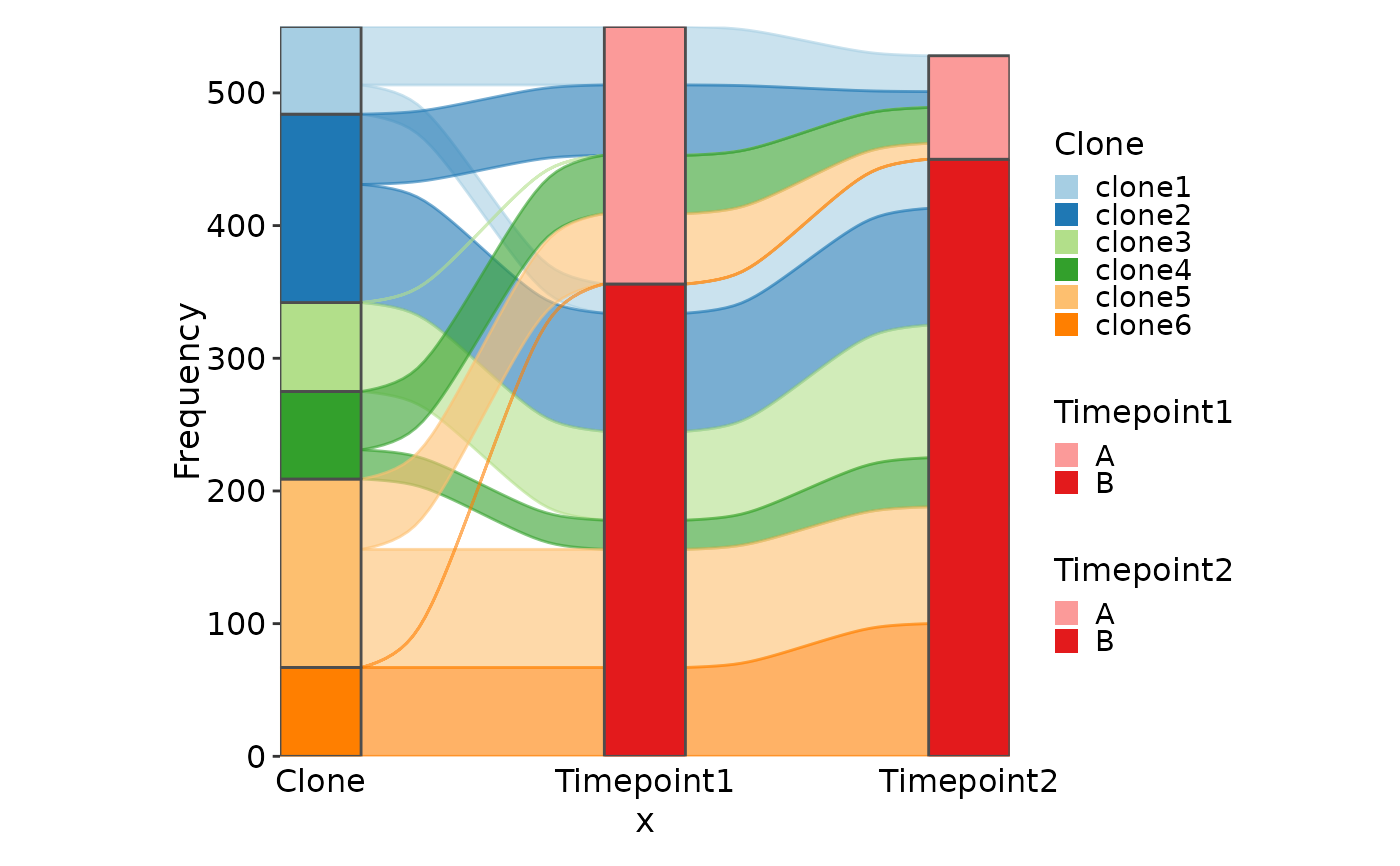
 df <- data.frame(
Clone = rep(paste0("clone", 1:6), each = 2),
Timepoint1 = sample(c(rep(0, 30), 1:100), 6),
Timepoint2 = sample(c(rep(0, 30), 1:100), 6),
Group = rep(c("A", "B"), 6)
)
SankeyPlot(df, x = c(".", "Timepoint1", "Timepoint2"),
stratum = "Group", links_fill_by = "Clone", links_color = ".fill")
df <- data.frame(
Clone = rep(paste0("clone", 1:6), each = 2),
Timepoint1 = sample(c(rep(0, 30), 1:100), 6),
Timepoint2 = sample(c(rep(0, 30), 1:100), 6),
Group = rep(c("A", "B"), 6)
)
SankeyPlot(df, x = c(".", "Timepoint1", "Timepoint2"),
stratum = "Group", links_fill_by = "Clone", links_color = ".fill")
 # }
# }
