This function creates a dimension reduction plot for a Seurat object
a Giotto object, a path to an .h5ad file or an opened H5File by hdf5r package.
It allows for various customizations such as grouping by metadata,
adding edges between cell neighbors, highlighting specific cells, and more.
This function is a wrapper around plotthis::DimPlot(), which provides a
flexible way to visualize cell clusters in reduced dimensions. This function
extracts the necessary data from the Seurat or Giotto object and passes it to
plotthis::DimPlot().
Usage
CellDimPlot(
object,
reduction = NULL,
graph = NULL,
group_by = NULL,
ident = NULL,
spat_unit = NULL,
feat_type = NULL,
velocity = NULL,
...
)Arguments
- object
A seurat object, a giotto object, a path to an .h5ad file or an opened
H5Filebyhdf5rpackage.- reduction
Name of the reduction to plot (for example, "umap").
- graph
Specify the graph name to add edges between cell neighbors to the plot.
- group_by
A character vector of column name(s) to group the data. Default is NULL.
- ident
Alias for
group_by.- spat_unit
The spatial unit to use for the plot. Only applied to Giotto objects.
- feat_type
feature type of the features (e.g. "rna", "dna", "protein"), only applied to Giotto objects.
- velocity
The name of velocity reduction to plot cell velocities. It is typically
"stochastic_<reduction>","deterministic_<reduction>", or"dynamical_<reduction>".- ...
Other arguments passed to
plotthis::DimPlot().
Details
See
https://pwwang.github.io/scplotter/articles/Giotto_CODEX.html
https://pwwang.github.io/scplotter/articles/Giotto_seqFISH.html
https://pwwang.github.io/scplotter/articles/Giotto_SlideSeq.html
https://pwwang.github.io/scplotter/articles/Giotto_Spatial_CITE-Seq.html
https://pwwang.github.io/scplotter/articles/Giotto_Visium.html
https://pwwang.github.io/scplotter/articles/Giotto_VisiumHD.html
https://pwwang.github.io/scplotter/articles/Giotto_Xenium.html
for examples of using this function with a Giotto object.
And see:
for examples of using this function with .h5ad files.
Examples
# \donttest{
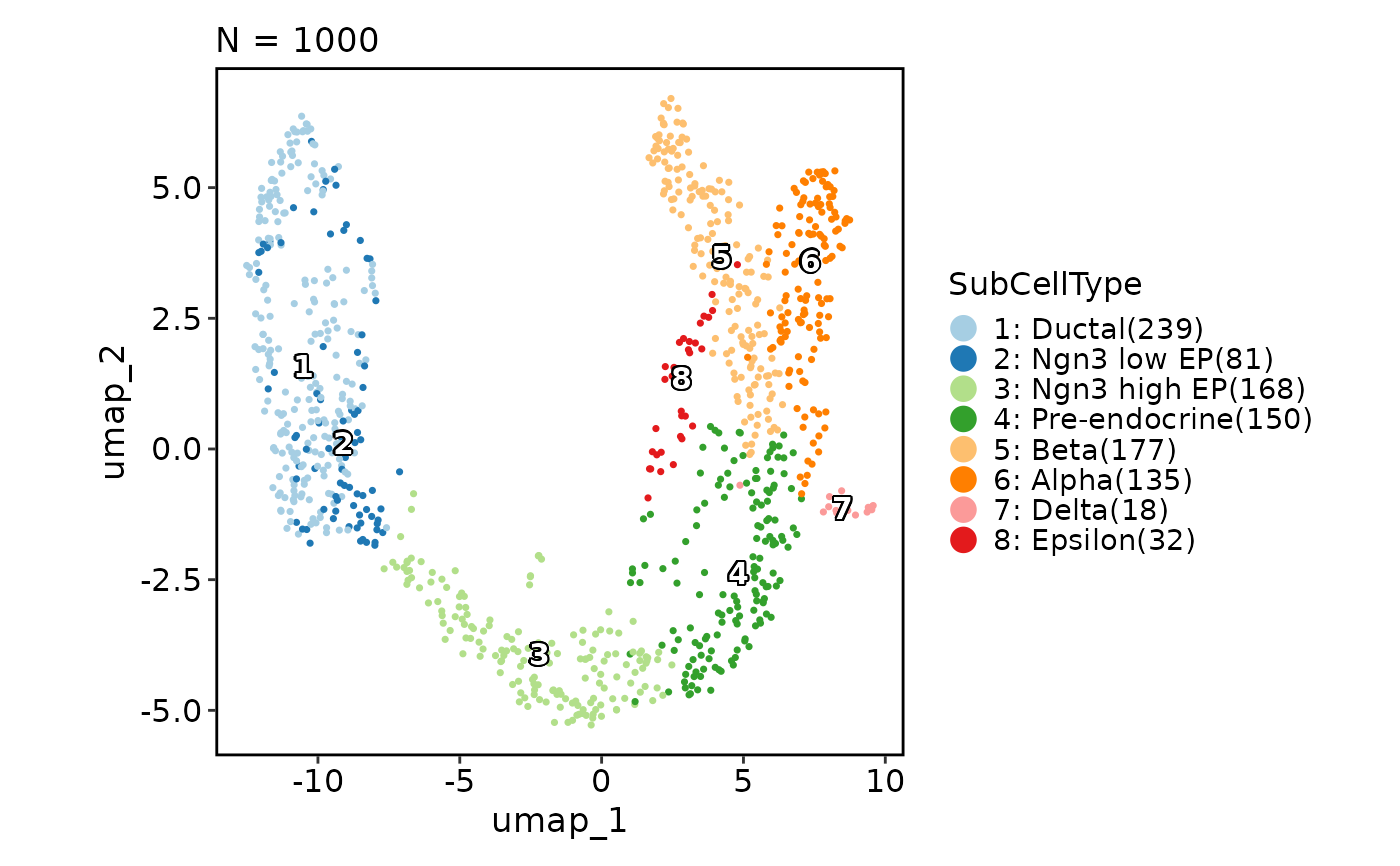
data(pancreas_sub)
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP")
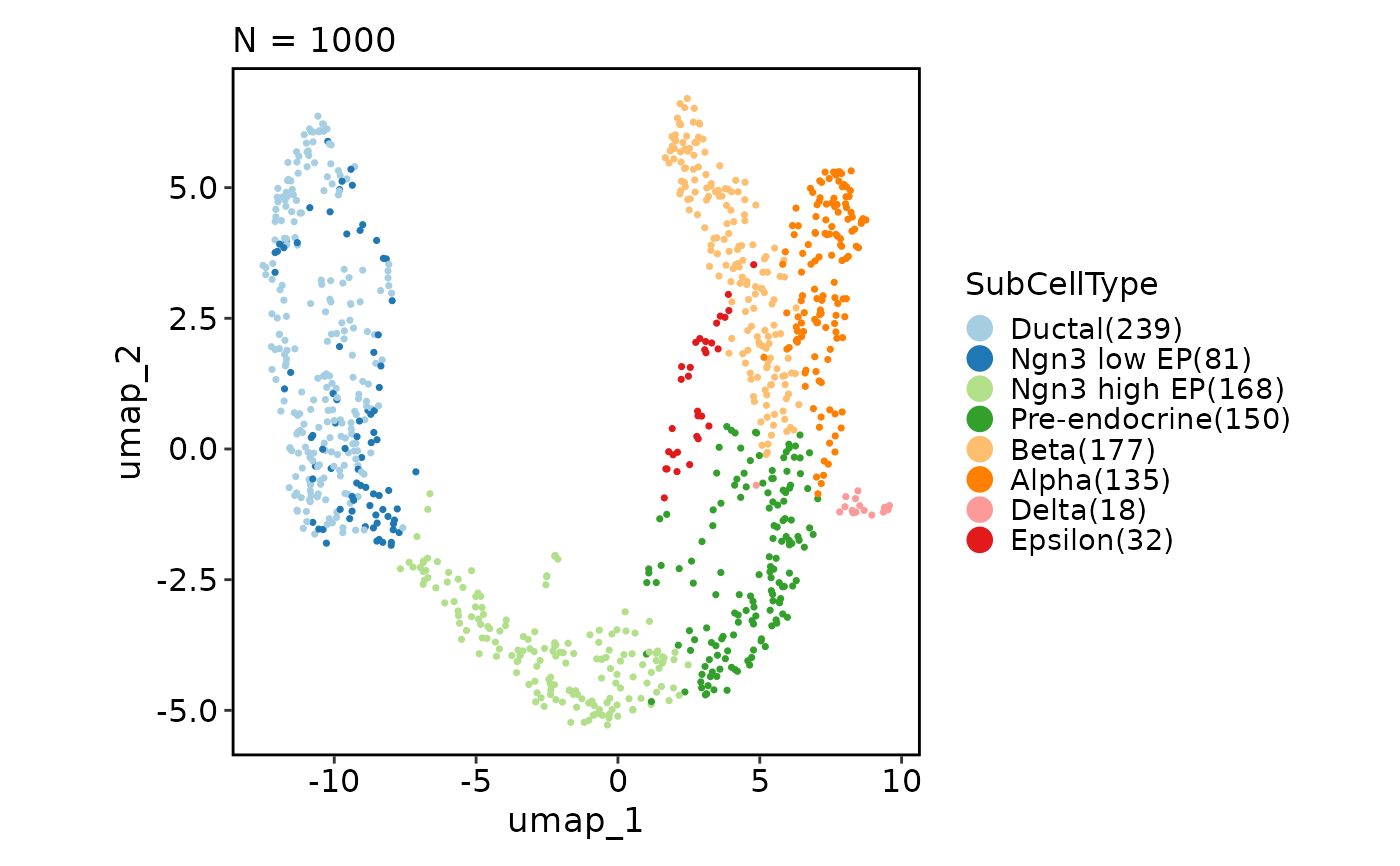 CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
theme = "theme_blank")
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
theme = "theme_blank")
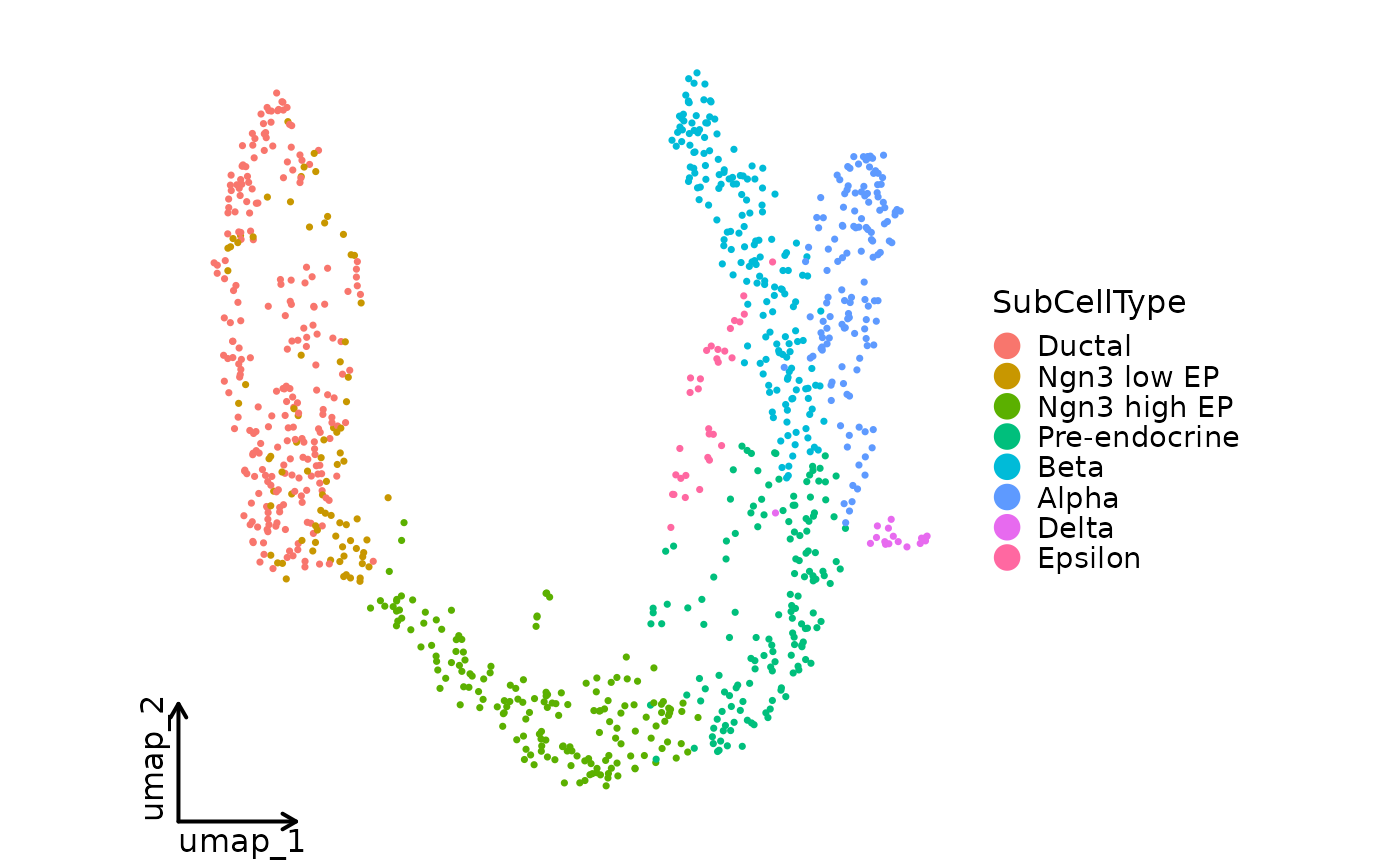
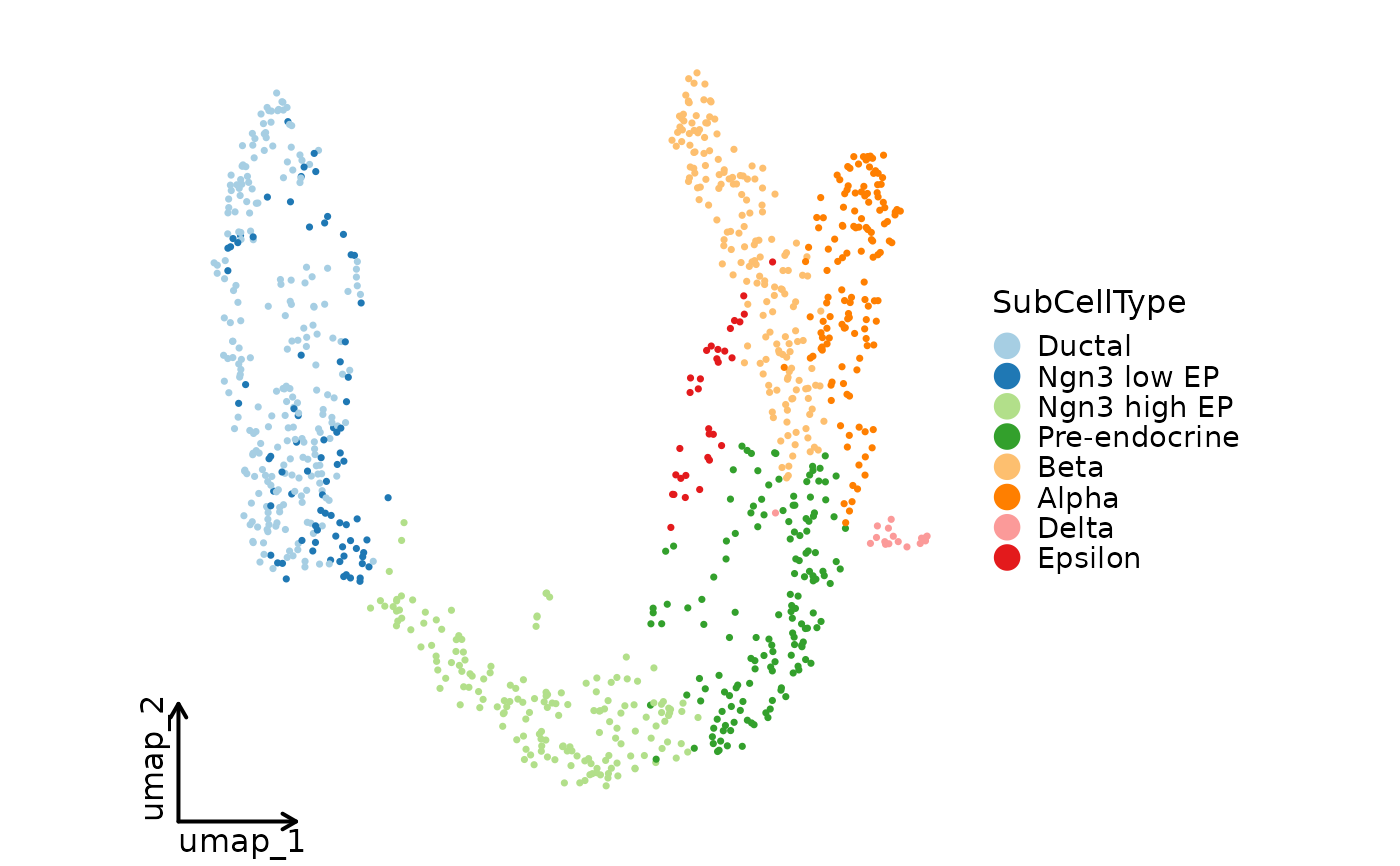 CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
palette = "seurat", theme = "theme_blank")
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
palette = "seurat", theme = "theme_blank")
 CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
theme = ggplot2::theme_classic, theme_args = list(base_size = 16))
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
theme = ggplot2::theme_classic, theme_args = list(base_size = 16))
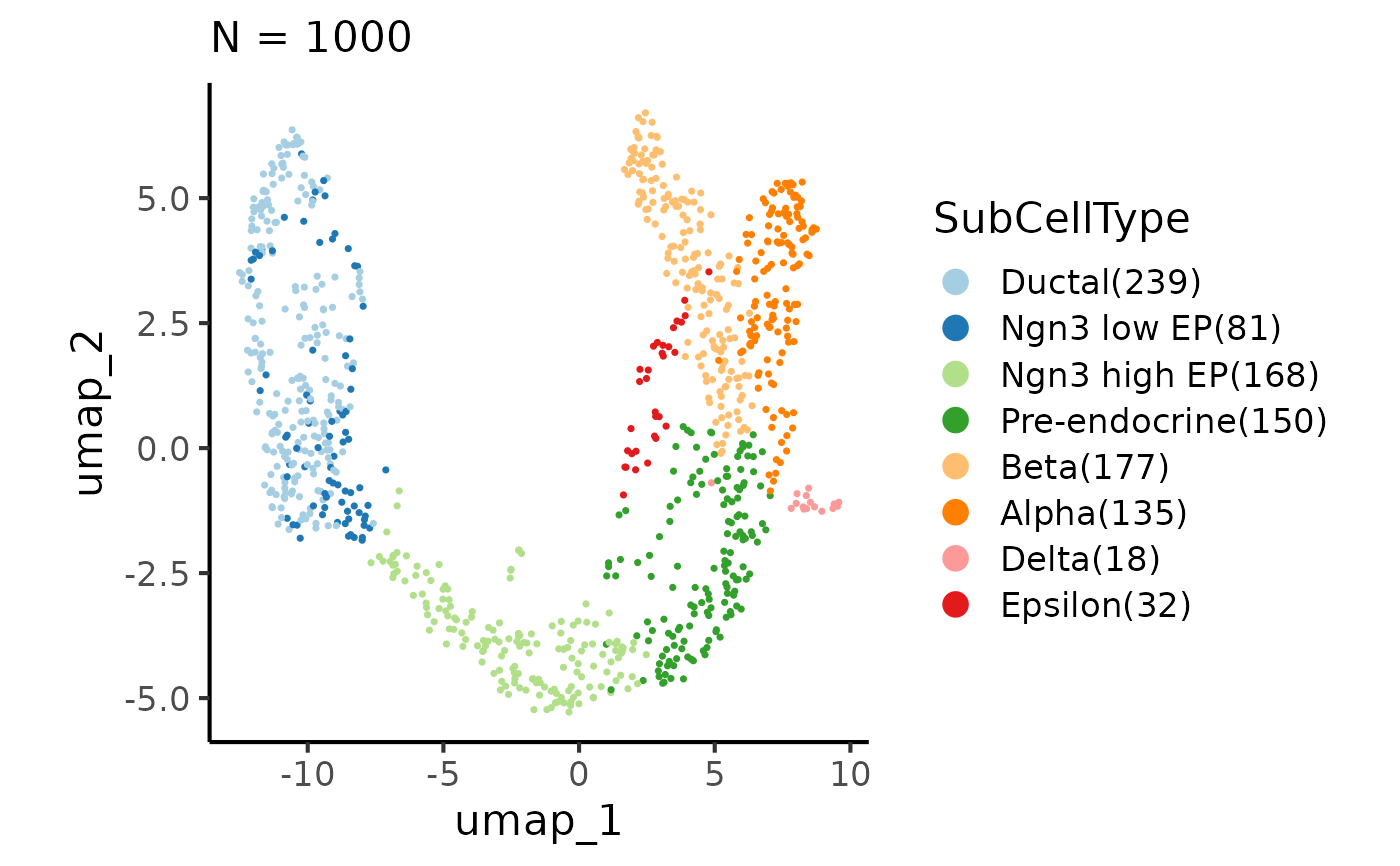 CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
raster = TRUE, raster_dpi = 30)
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
raster = TRUE, raster_dpi = 30)
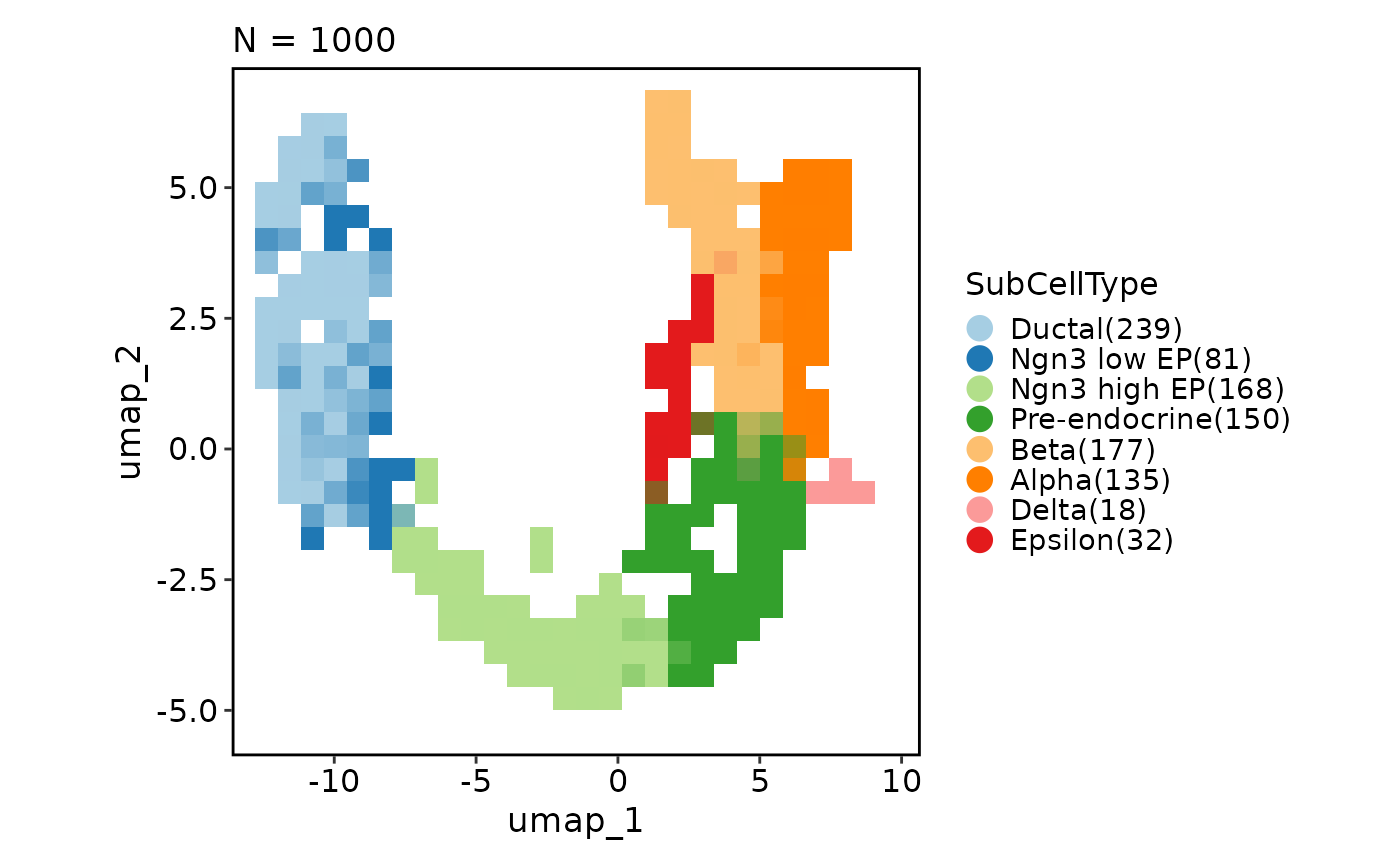 # Highlight cells
CellDimPlot(pancreas_sub,
group_by = "SubCellType", reduction = "UMAP",
highlight = 'SubCellType == "Epsilon"'
)
# Highlight cells
CellDimPlot(pancreas_sub,
group_by = "SubCellType", reduction = "UMAP",
highlight = 'SubCellType == "Epsilon"'
)
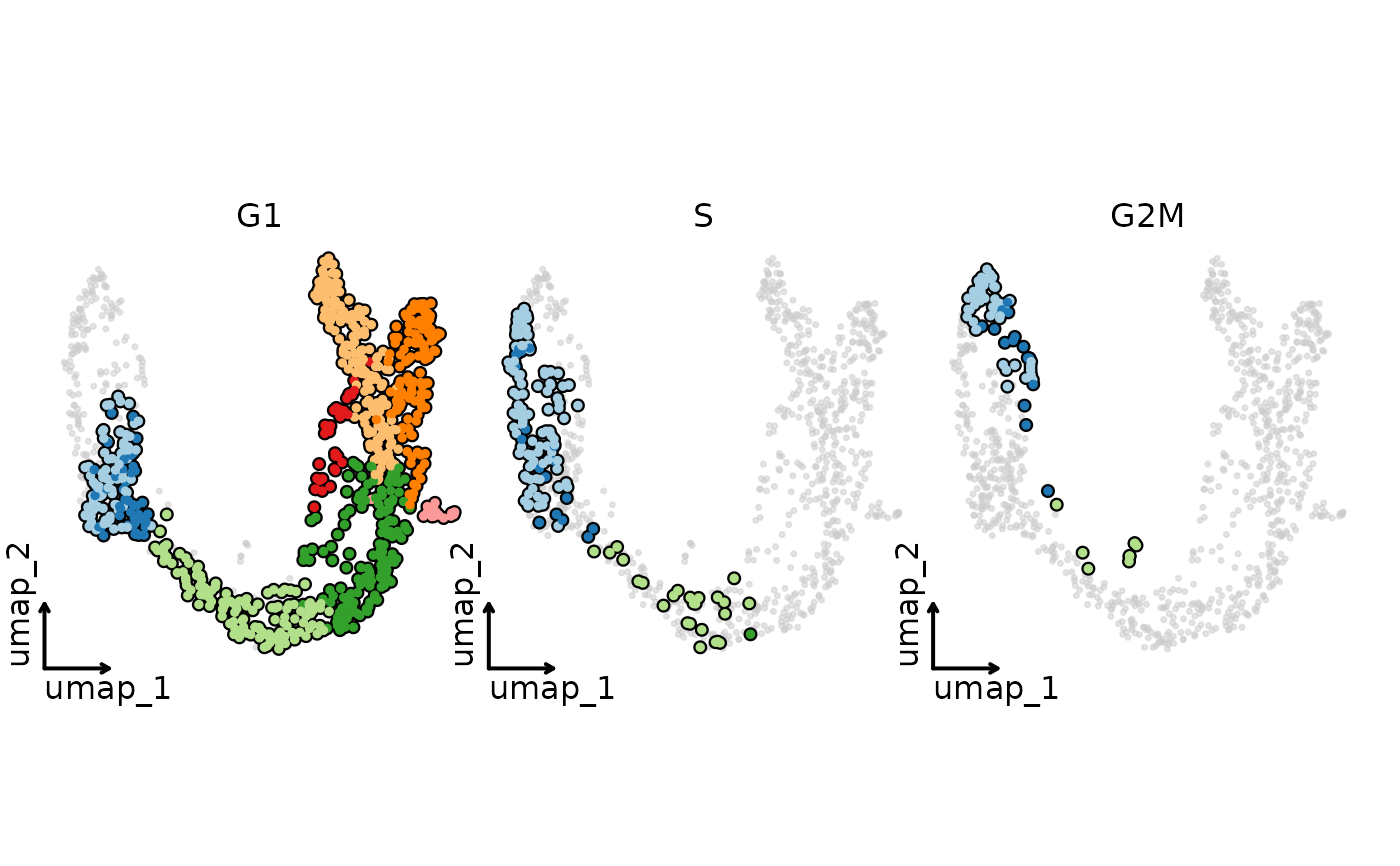
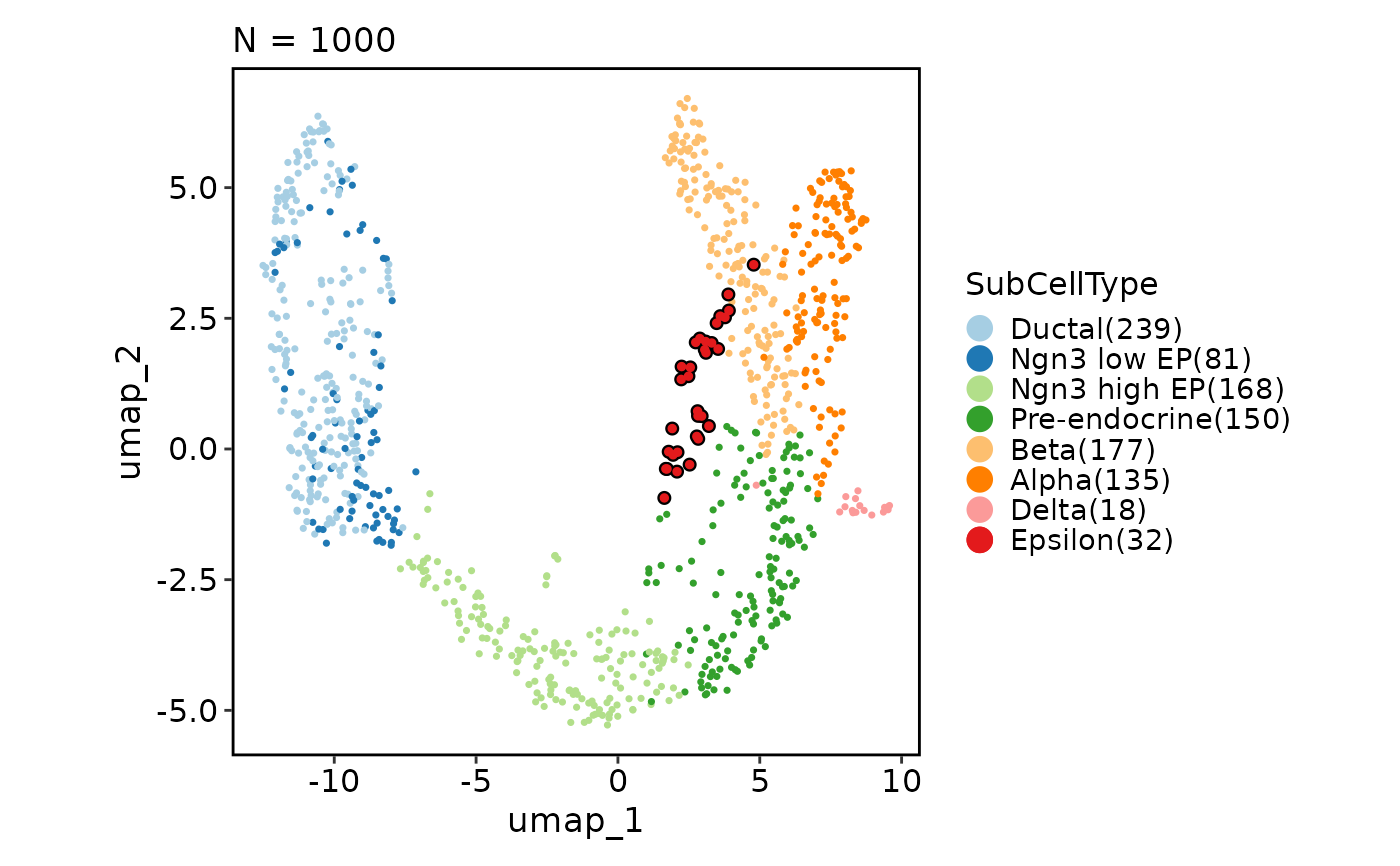 CellDimPlot(pancreas_sub,
group_by = "SubCellType", split_by = "Phase", reduction = "UMAP",
highlight = TRUE, theme = "theme_blank", legend.position = "none"
)
CellDimPlot(pancreas_sub,
group_by = "SubCellType", split_by = "Phase", reduction = "UMAP",
highlight = TRUE, theme = "theme_blank", legend.position = "none"
)
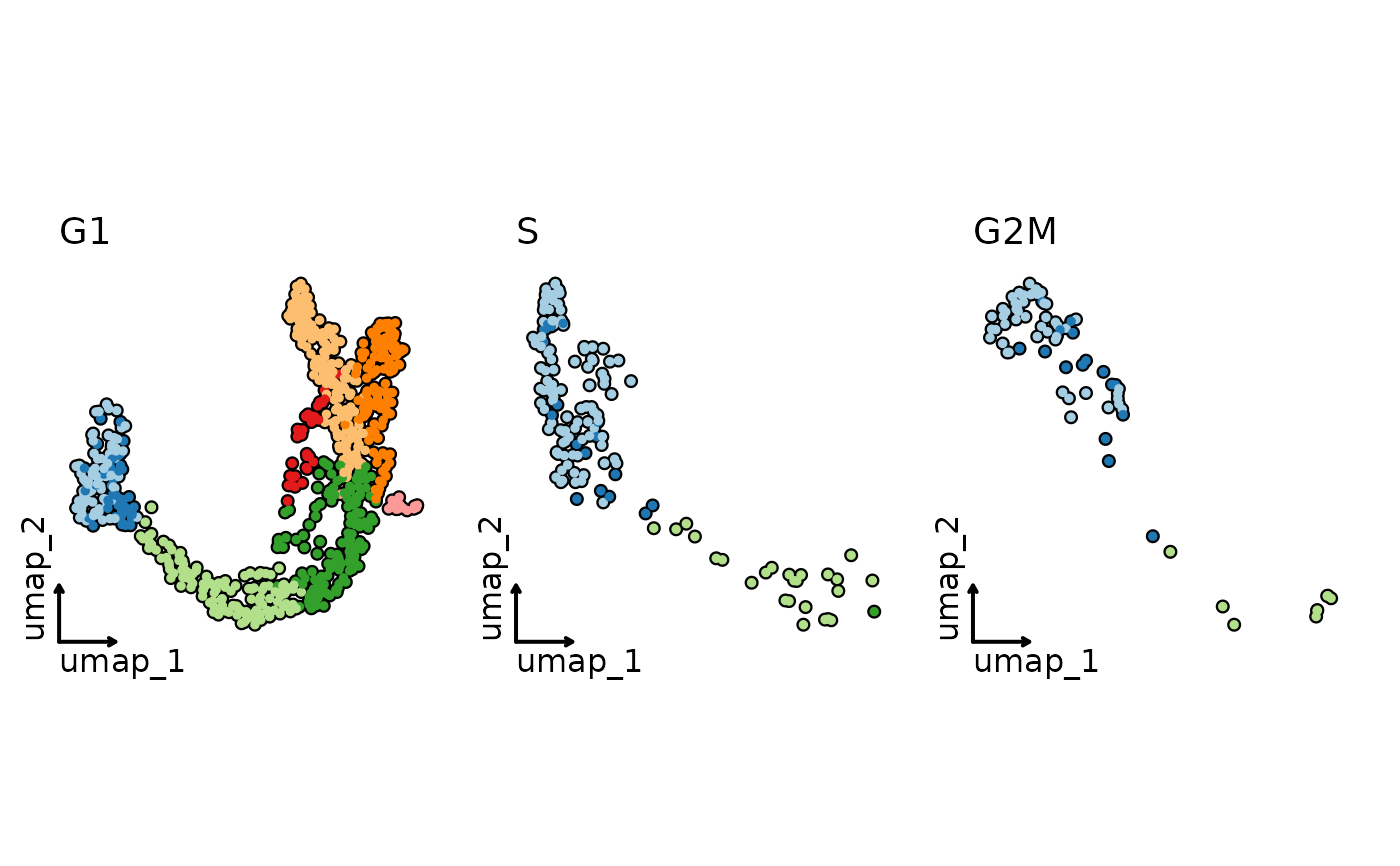 CellDimPlot(pancreas_sub,
group_by = "SubCellType", facet_by = "Phase", reduction = "UMAP",
highlight = TRUE, theme = "theme_blank", legend.position = "none"
)
CellDimPlot(pancreas_sub,
group_by = "SubCellType", facet_by = "Phase", reduction = "UMAP",
highlight = TRUE, theme = "theme_blank", legend.position = "none"
)
 # Add group labels
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
label = TRUE)
# Add group labels
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
label = TRUE)
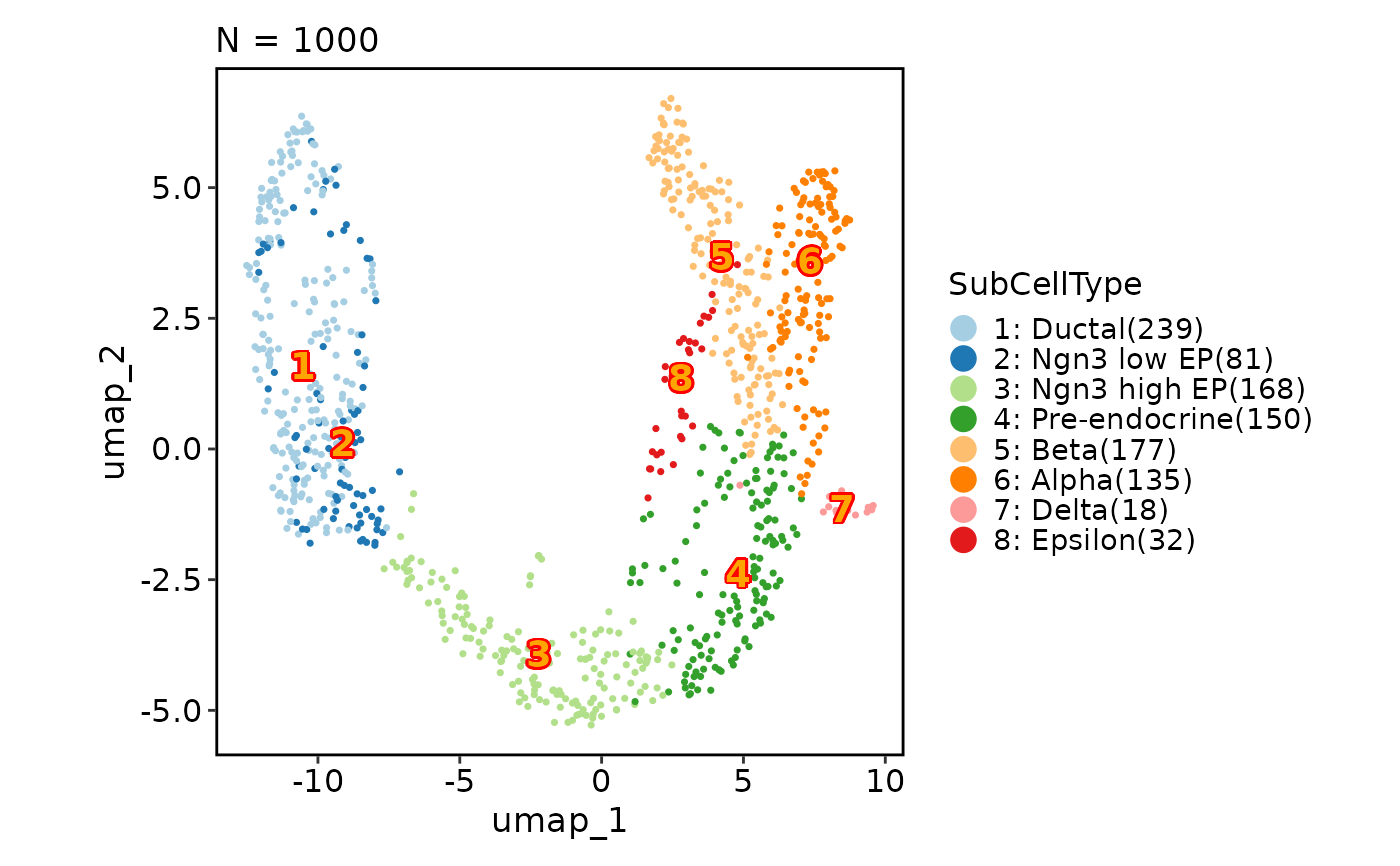
 CellDimPlot(pancreas_sub,
group_by = "SubCellType", reduction = "UMAP",
label = TRUE, label_fg = "orange", label_bg = "red", label_size = 5
)
CellDimPlot(pancreas_sub,
group_by = "SubCellType", reduction = "UMAP",
label = TRUE, label_fg = "orange", label_bg = "red", label_size = 5
)
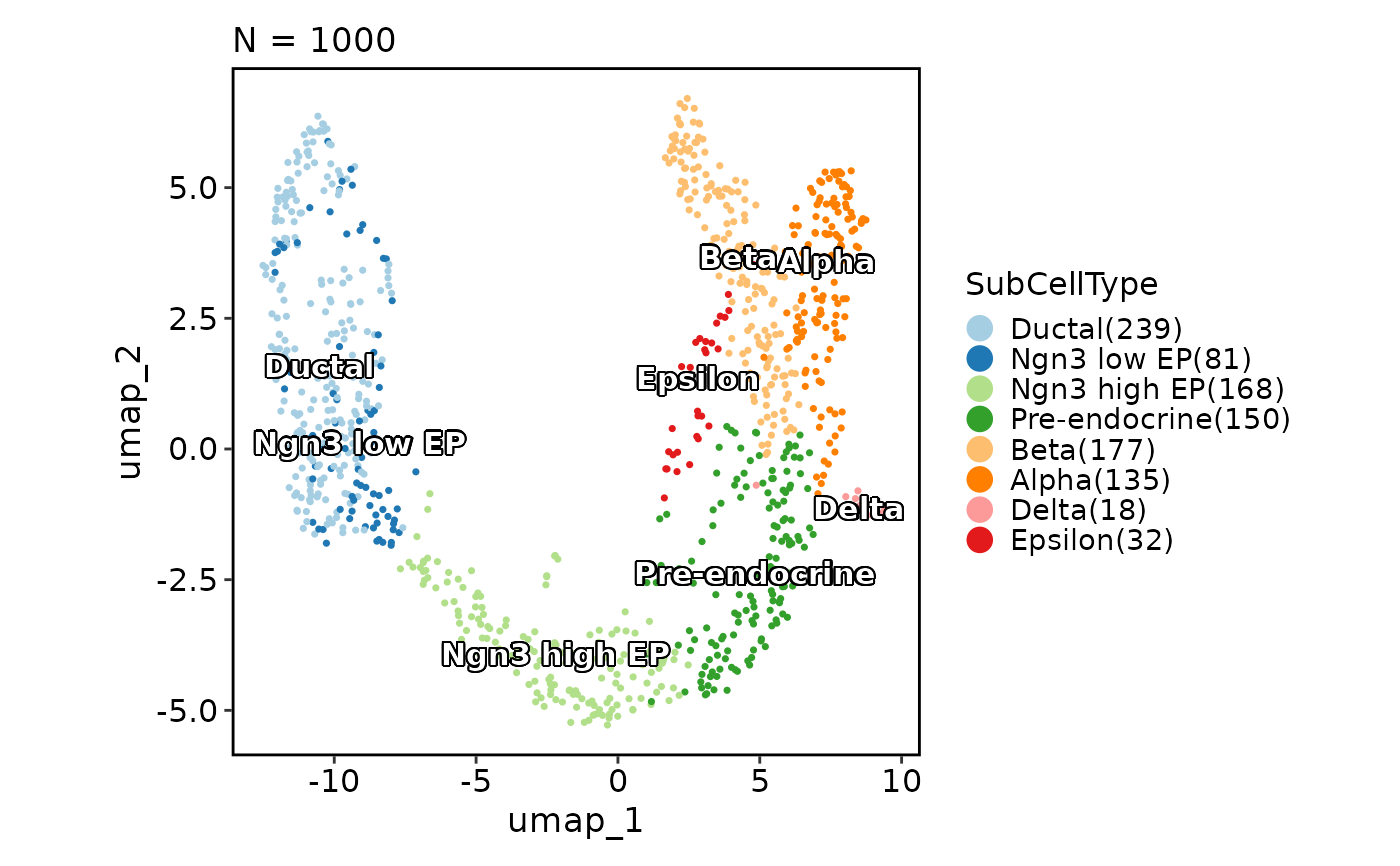
 CellDimPlot(pancreas_sub,
group_by = "SubCellType", reduction = "UMAP",
label = TRUE, label_insitu = TRUE
)
CellDimPlot(pancreas_sub,
group_by = "SubCellType", reduction = "UMAP",
label = TRUE, label_insitu = TRUE
)
 CellDimPlot(pancreas_sub,
group_by = "SubCellType", reduction = "UMAP",
label = TRUE, label_insitu = TRUE, label_repel = TRUE,
label_segment_color = "red"
)
CellDimPlot(pancreas_sub,
group_by = "SubCellType", reduction = "UMAP",
label = TRUE, label_insitu = TRUE, label_repel = TRUE,
label_segment_color = "red"
)
 # Add various shape of marks
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
add_mark = TRUE)
# Add various shape of marks
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
add_mark = TRUE)
 CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
add_mark = TRUE, mark_expand = grid::unit(1, "mm"))
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
add_mark = TRUE, mark_expand = grid::unit(1, "mm"))
 CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
add_mark = TRUE, mark_alpha = 0.3)
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
add_mark = TRUE, mark_alpha = 0.3)
 CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
add_mark = TRUE, mark_linetype = 2)
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
add_mark = TRUE, mark_linetype = 2)
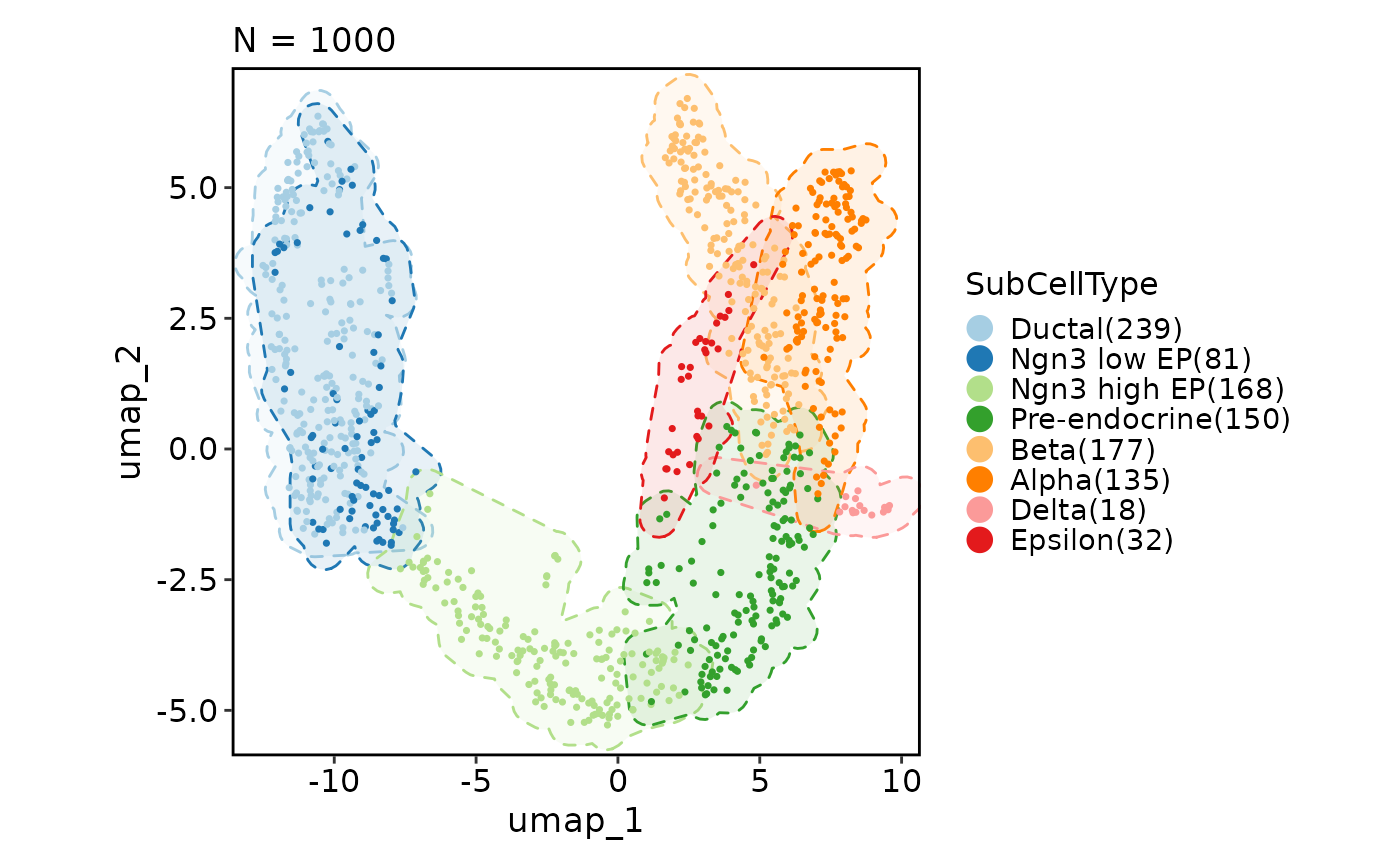 CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
add_mark = TRUE, mark_type = "ellipse")
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
add_mark = TRUE, mark_type = "ellipse")
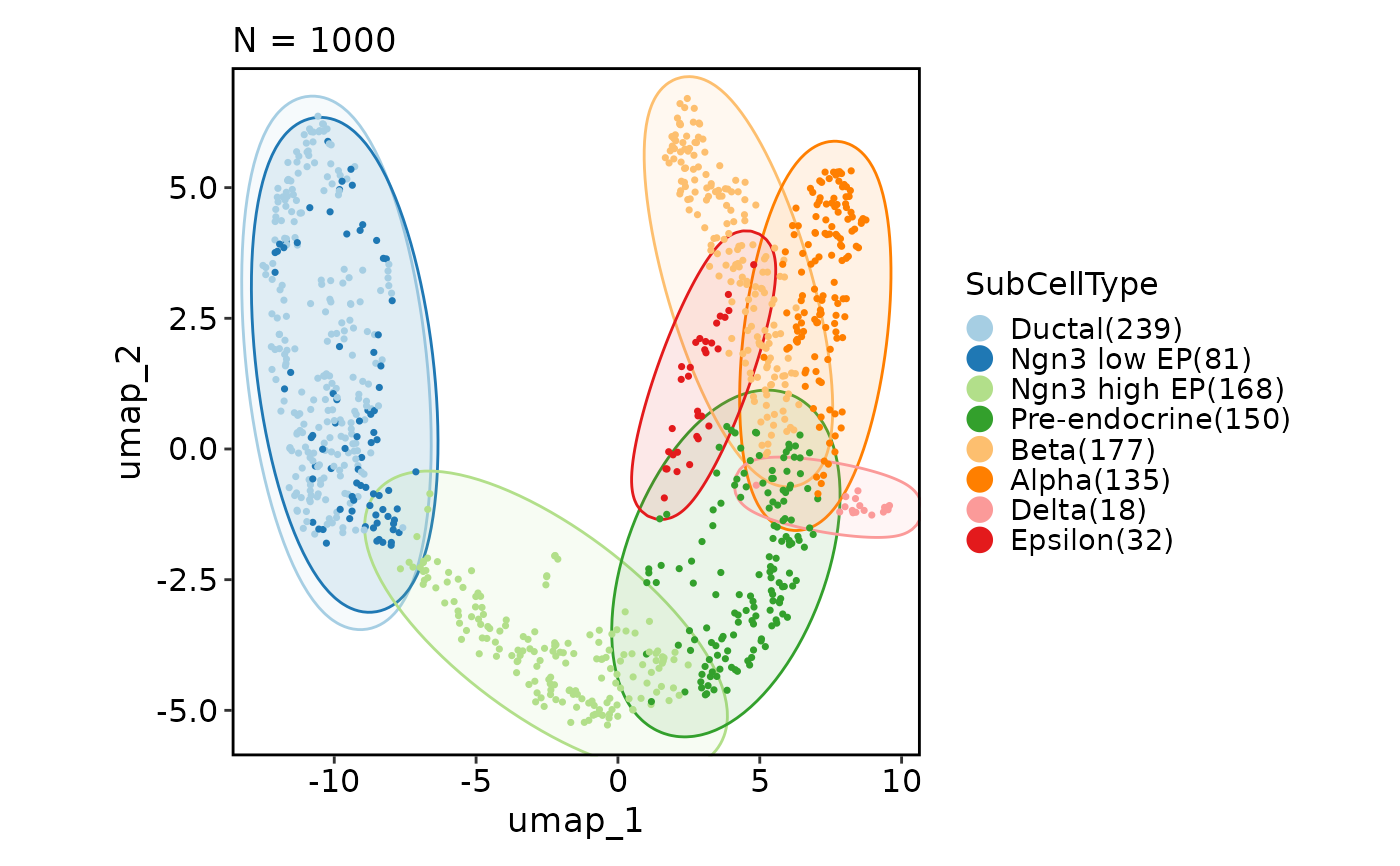 CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
add_mark = TRUE, mark_type = "rect")
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
add_mark = TRUE, mark_type = "rect")
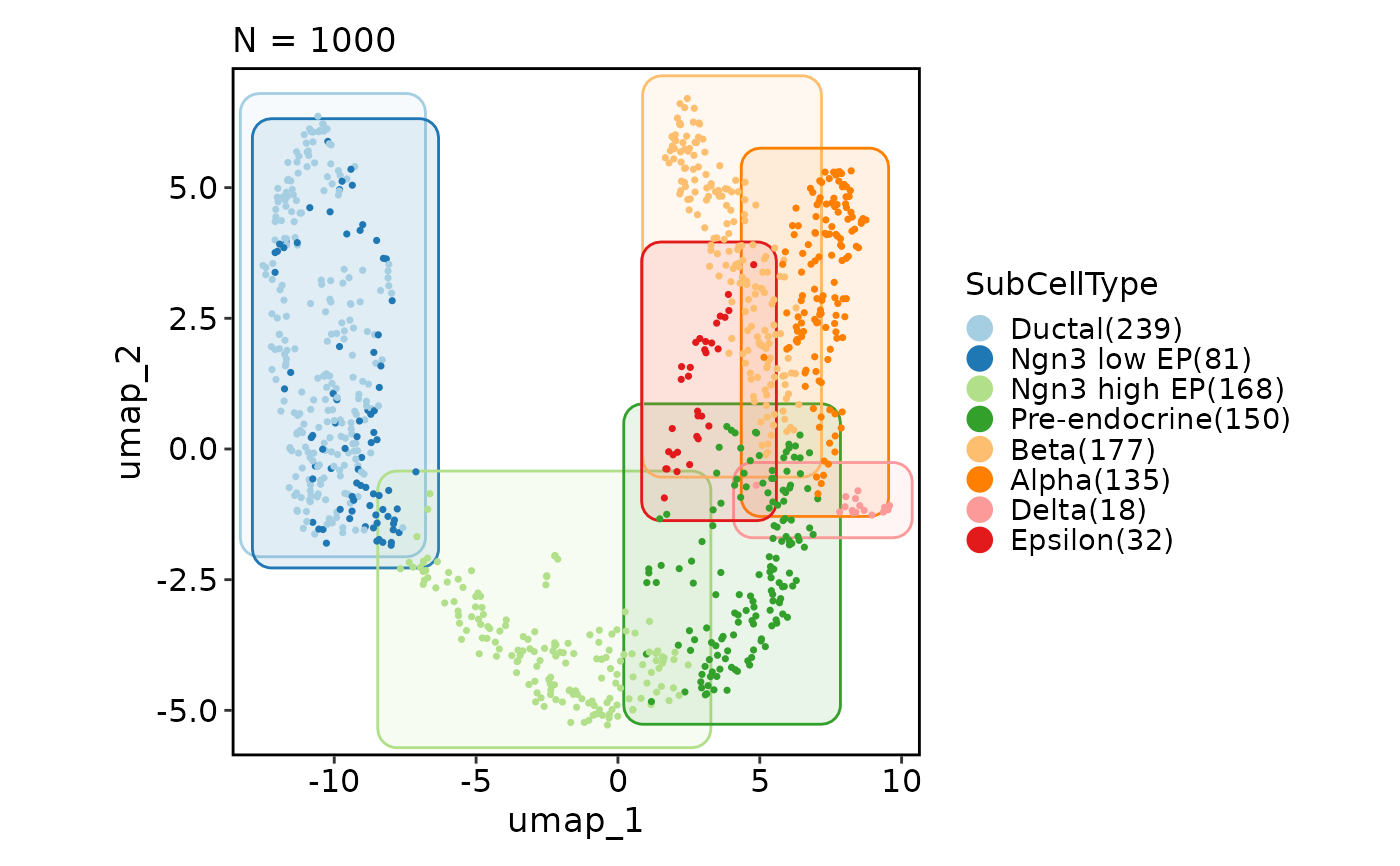 CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
add_mark = TRUE, mark_type = "circle")
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
add_mark = TRUE, mark_type = "circle")
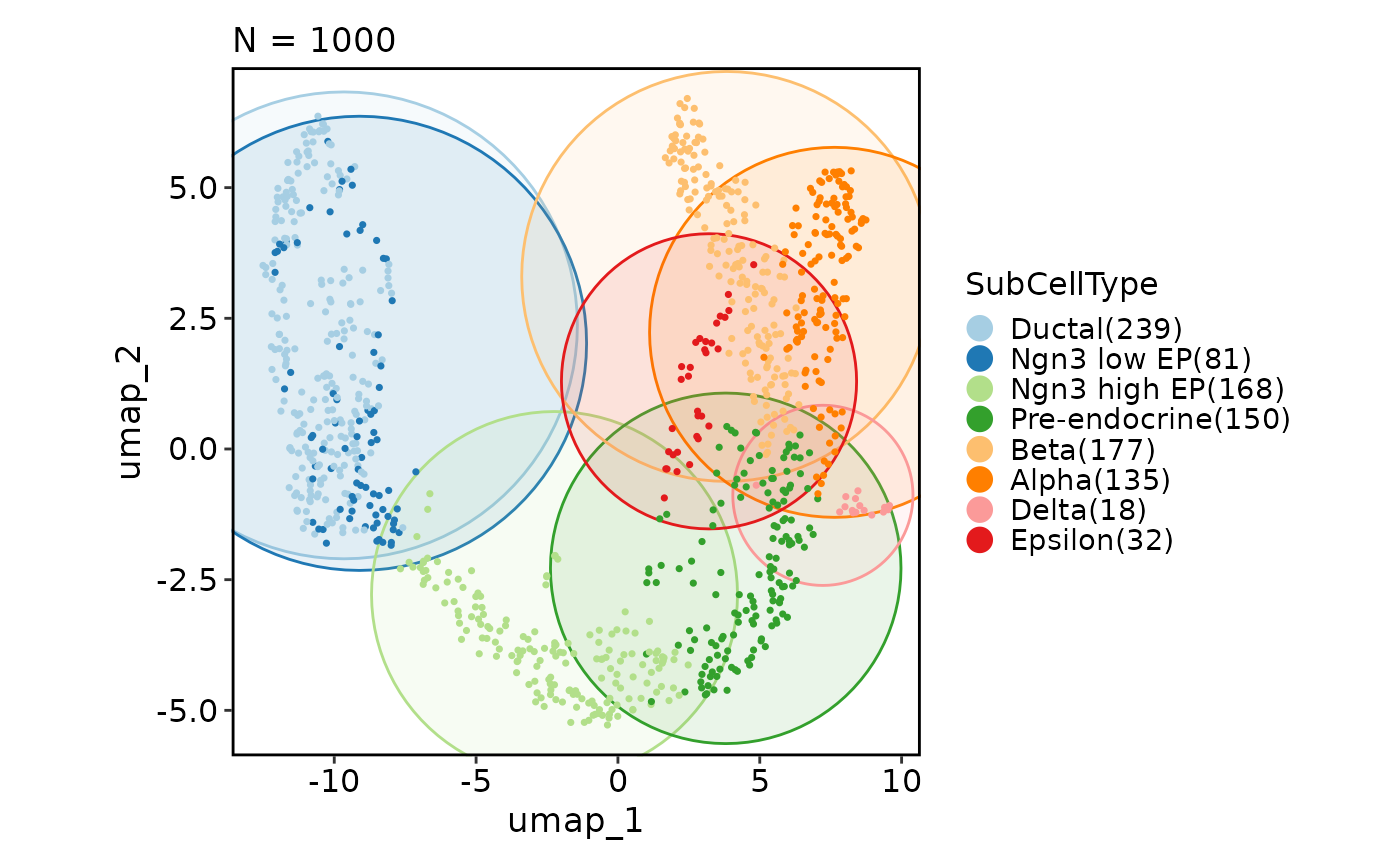 # Add a density layer
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
add_density = TRUE)
# Add a density layer
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
add_density = TRUE)
 CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
add_density = TRUE, density_filled = TRUE)
#> Warning: Removed 396 rows containing missing values or values outside the scale range
#> (`geom_raster()`).
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
add_density = TRUE, density_filled = TRUE)
#> Warning: Removed 396 rows containing missing values or values outside the scale range
#> (`geom_raster()`).
 CellDimPlot(pancreas_sub,
group_by = "SubCellType", reduction = "UMAP",
add_density = TRUE, density_filled = TRUE, density_filled_palette = "Blues",
highlight = TRUE
)
#> Warning: Removed 396 rows containing missing values or values outside the scale range
#> (`geom_raster()`).
CellDimPlot(pancreas_sub,
group_by = "SubCellType", reduction = "UMAP",
add_density = TRUE, density_filled = TRUE, density_filled_palette = "Blues",
highlight = TRUE
)
#> Warning: Removed 396 rows containing missing values or values outside the scale range
#> (`geom_raster()`).
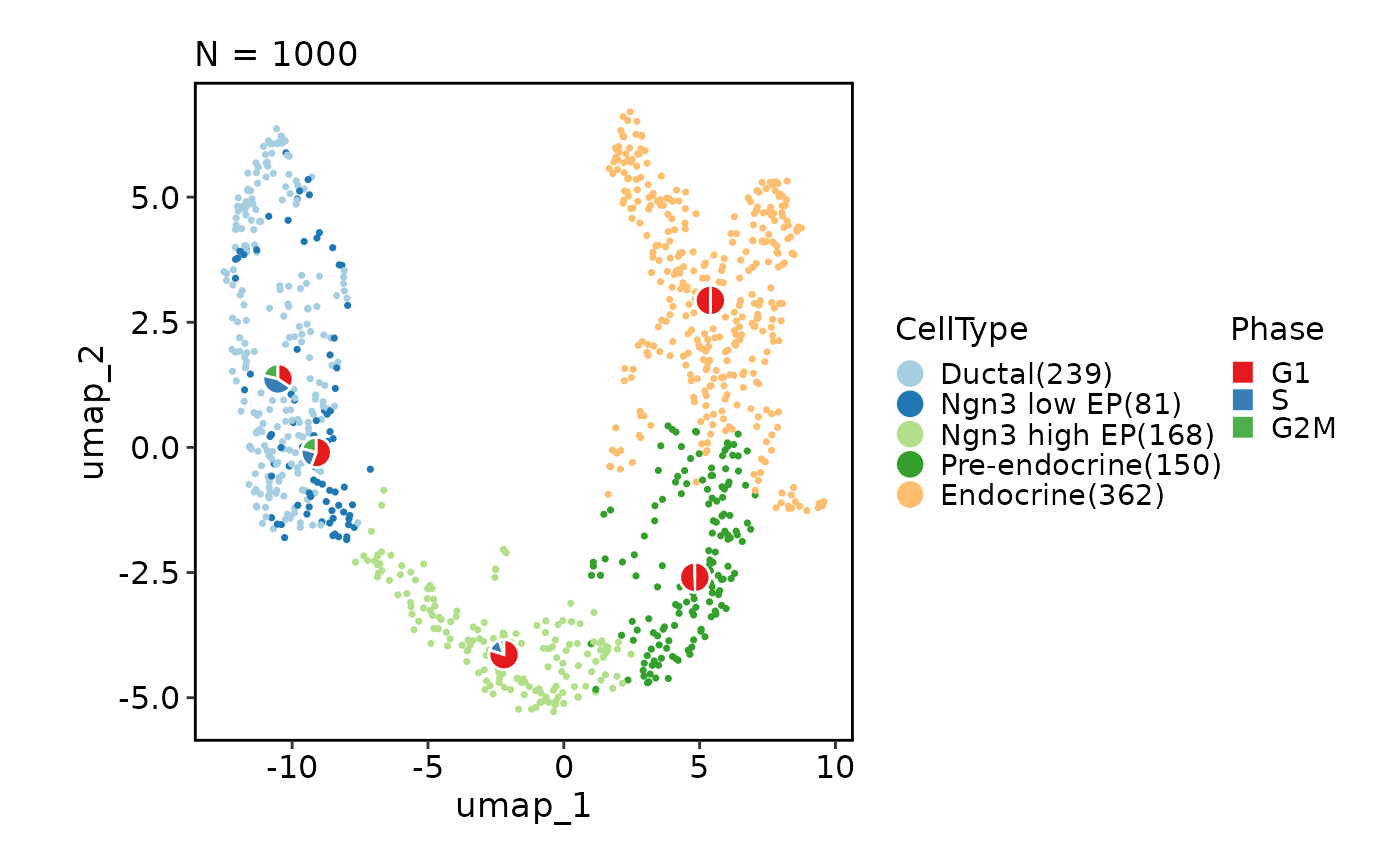
 # Add statistical charts
CellDimPlot(pancreas_sub,
group_by = "CellType", reduction = "UMAP", stat_by = "Phase")
# Add statistical charts
CellDimPlot(pancreas_sub,
group_by = "CellType", reduction = "UMAP", stat_by = "Phase")
 CellDimPlot(pancreas_sub,
group_by = "CellType", reduction = "UMAP", stat_by = "Phase",
stat_plot_type = "ring", stat_plot_label = TRUE, stat_plot_size = 0.15)
CellDimPlot(pancreas_sub,
group_by = "CellType", reduction = "UMAP", stat_by = "Phase",
stat_plot_type = "ring", stat_plot_label = TRUE, stat_plot_size = 0.15)
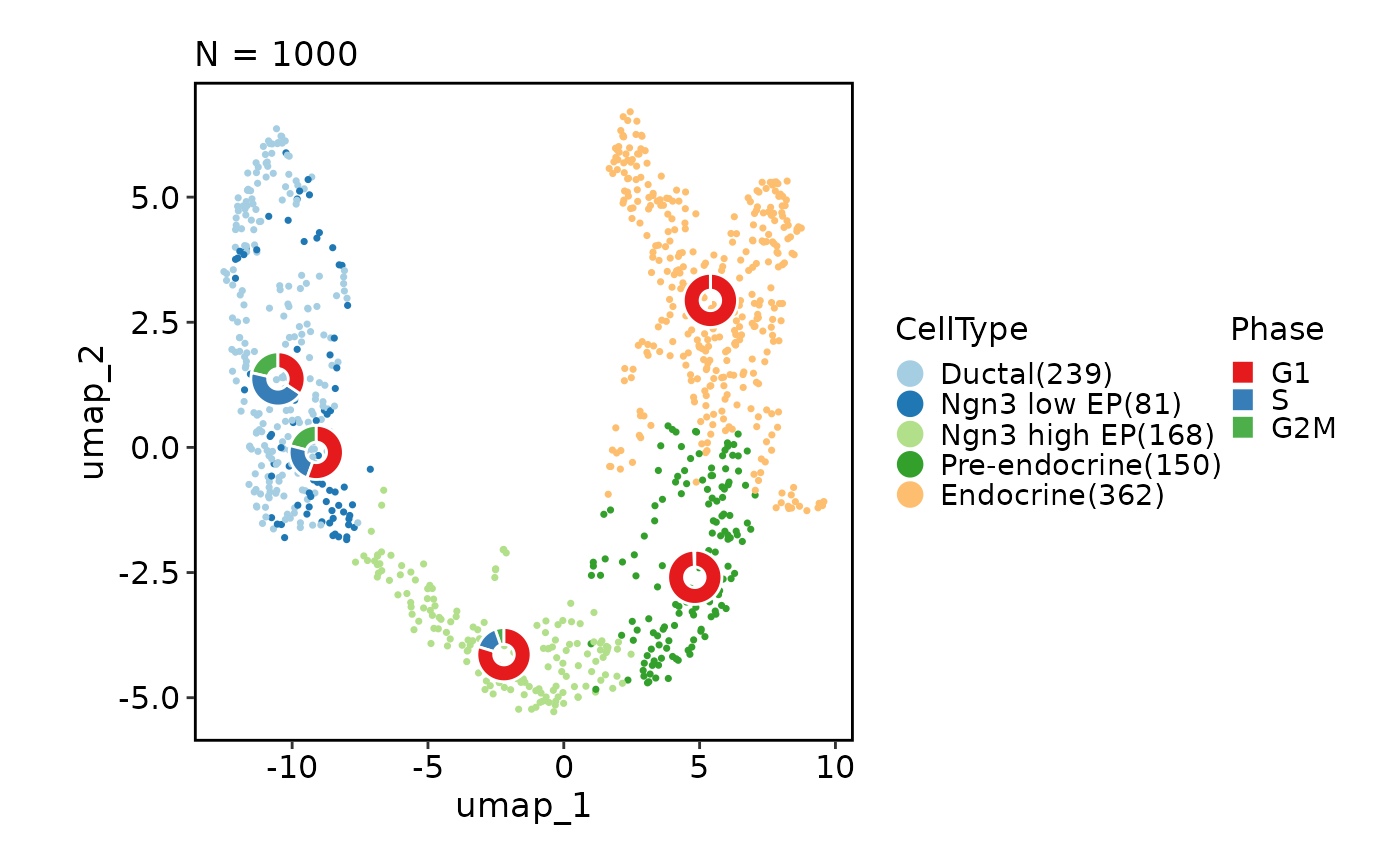 CellDimPlot(pancreas_sub,
group_by = "CellType", reduction = "UMAP", stat_by = "Phase",
stat_plot_type = "bar", stat_type = "count")
CellDimPlot(pancreas_sub,
group_by = "CellType", reduction = "UMAP", stat_by = "Phase",
stat_plot_type = "bar", stat_type = "count")
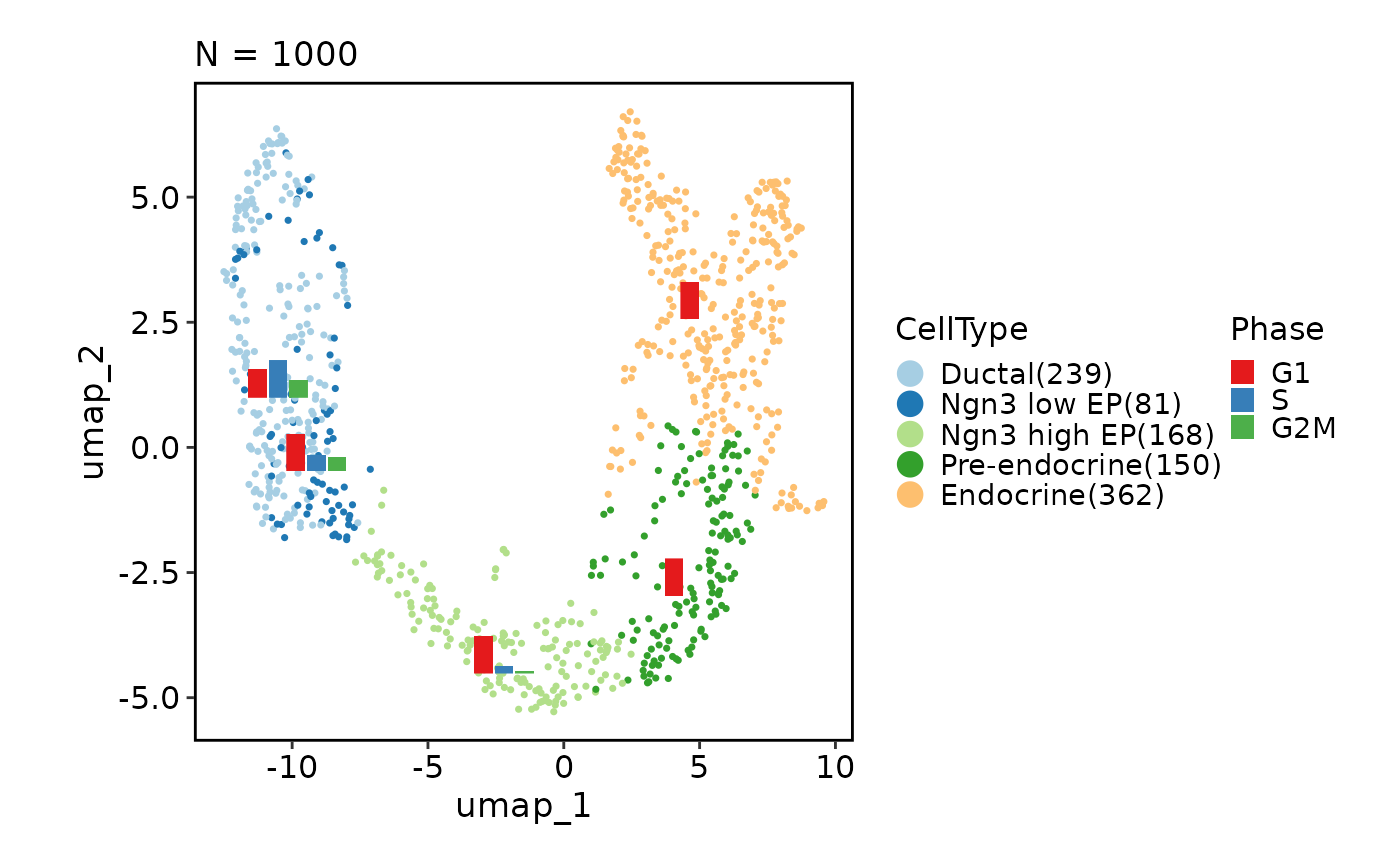 CellDimPlot(pancreas_sub,
group_by = "CellType", reduction = "UMAP", stat_by = "Phase",
stat_plot_type = "line", stat_type = "count", stat_args = list(point_size = 1))
CellDimPlot(pancreas_sub,
group_by = "CellType", reduction = "UMAP", stat_by = "Phase",
stat_plot_type = "line", stat_type = "count", stat_args = list(point_size = 1))
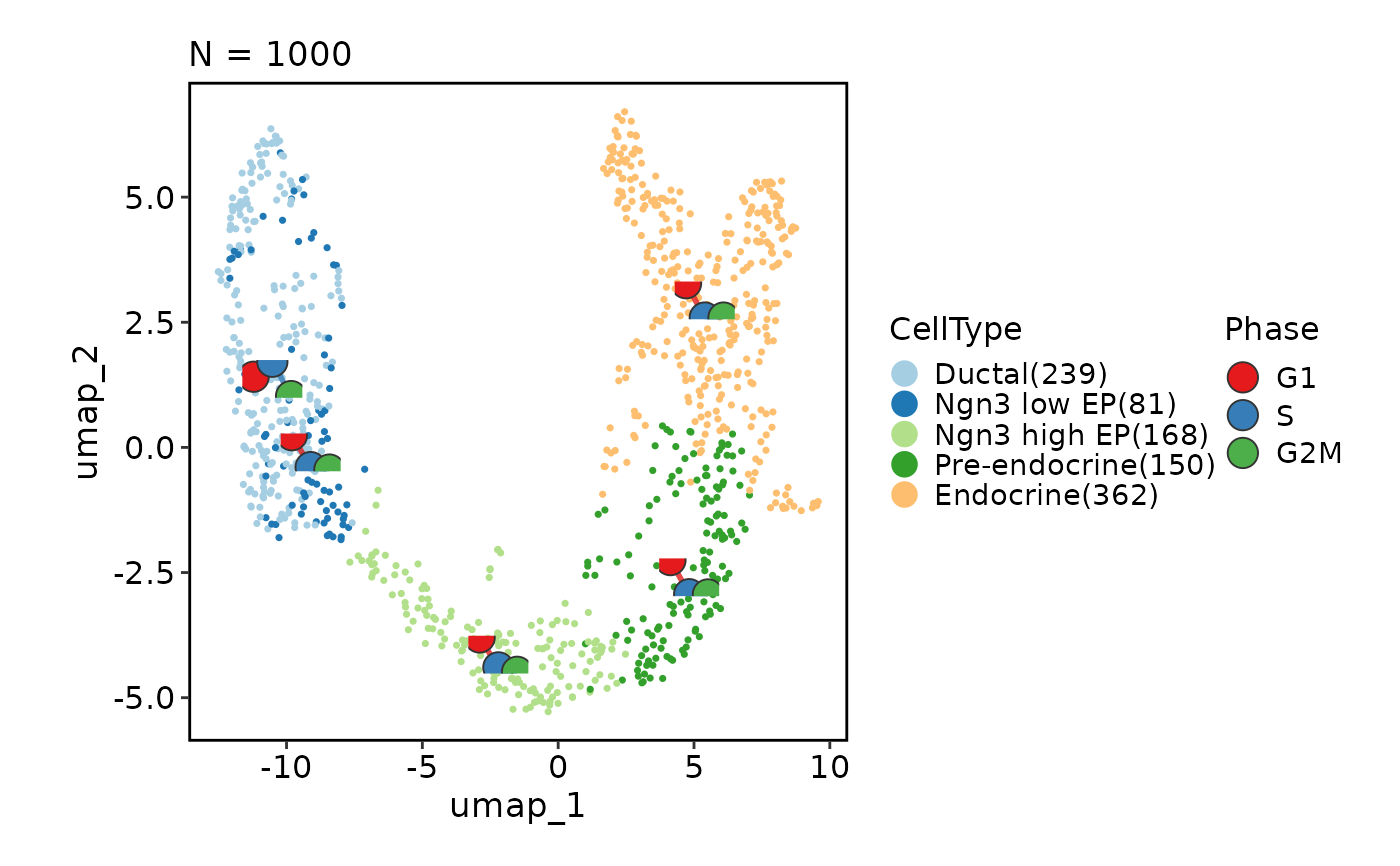 # Chane the plot type from point to the hexagonal bin
CellDimPlot(pancreas_sub, group_by = "CellType", reduction = "UMAP",
hex = TRUE)
#> Warning: Removed 5 rows containing missing values or values outside the scale range
#> (`geom_hex()`).
# Chane the plot type from point to the hexagonal bin
CellDimPlot(pancreas_sub, group_by = "CellType", reduction = "UMAP",
hex = TRUE)
#> Warning: Removed 5 rows containing missing values or values outside the scale range
#> (`geom_hex()`).
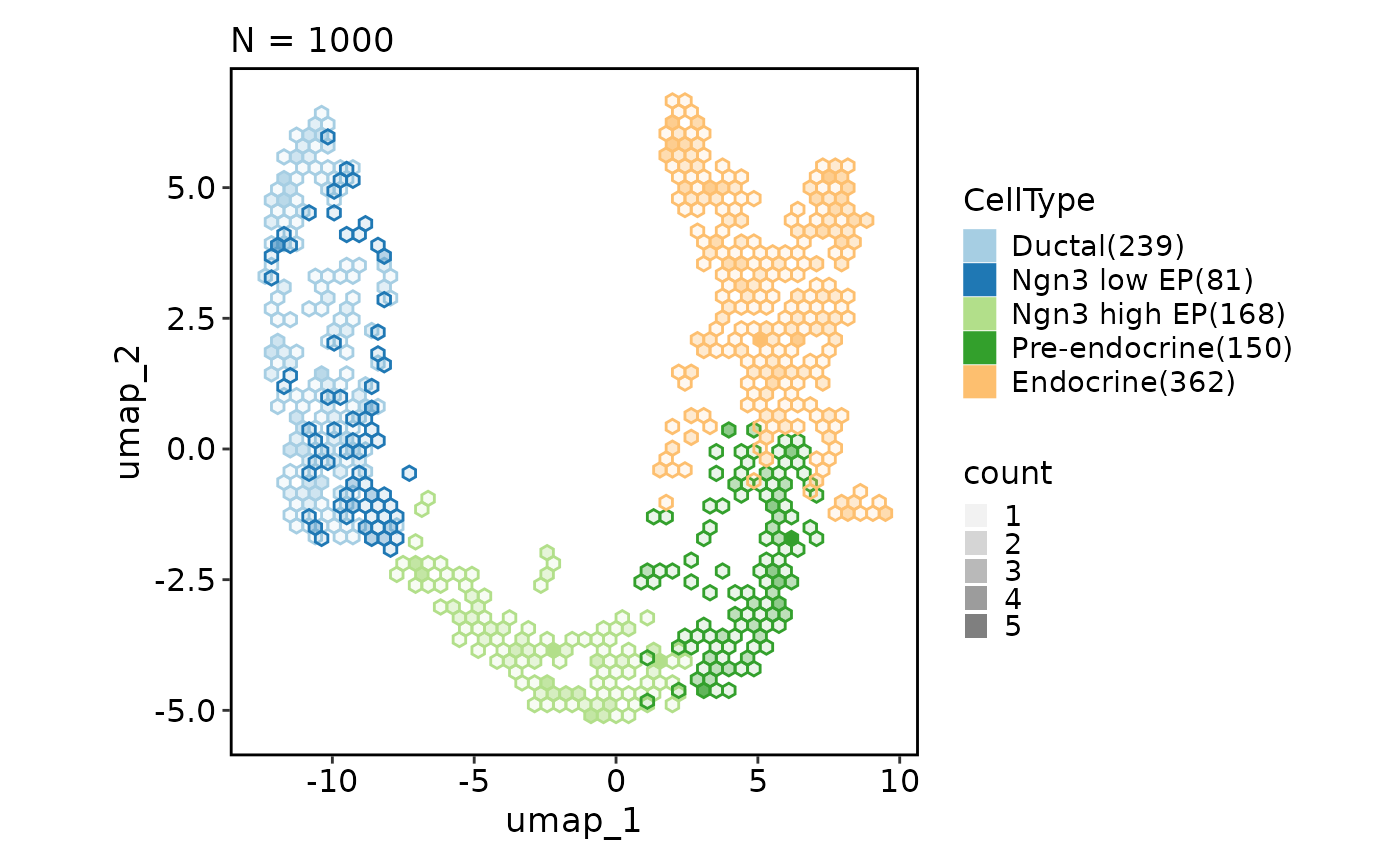 CellDimPlot(pancreas_sub, group_by = "CellType", reduction = "UMAP",
hex = TRUE, hex_bins = 20)
#> Warning: Removed 4 rows containing missing values or values outside the scale range
#> (`geom_hex()`).
CellDimPlot(pancreas_sub, group_by = "CellType", reduction = "UMAP",
hex = TRUE, hex_bins = 20)
#> Warning: Removed 4 rows containing missing values or values outside the scale range
#> (`geom_hex()`).
 CellDimPlot(pancreas_sub, group_by = "CellType", reduction = "UMAP",
hex = TRUE, hex_count = FALSE)
#> Warning: Removed 5 rows containing missing values or values outside the scale range
#> (`geom_hex()`).
CellDimPlot(pancreas_sub, group_by = "CellType", reduction = "UMAP",
hex = TRUE, hex_count = FALSE)
#> Warning: Removed 5 rows containing missing values or values outside the scale range
#> (`geom_hex()`).
 # Show neighbors graphs on the plot
CellDimPlot(pancreas_sub, group_by = "CellType", reduction = "UMAP",
graph = "RNA_nn")
# Show neighbors graphs on the plot
CellDimPlot(pancreas_sub, group_by = "CellType", reduction = "UMAP",
graph = "RNA_nn")
 CellDimPlot(pancreas_sub, group_by = "CellType", reduction = "UMAP",
graph = "RNA_snn", edge_color = "grey80")
CellDimPlot(pancreas_sub, group_by = "CellType", reduction = "UMAP",
graph = "RNA_snn", edge_color = "grey80")
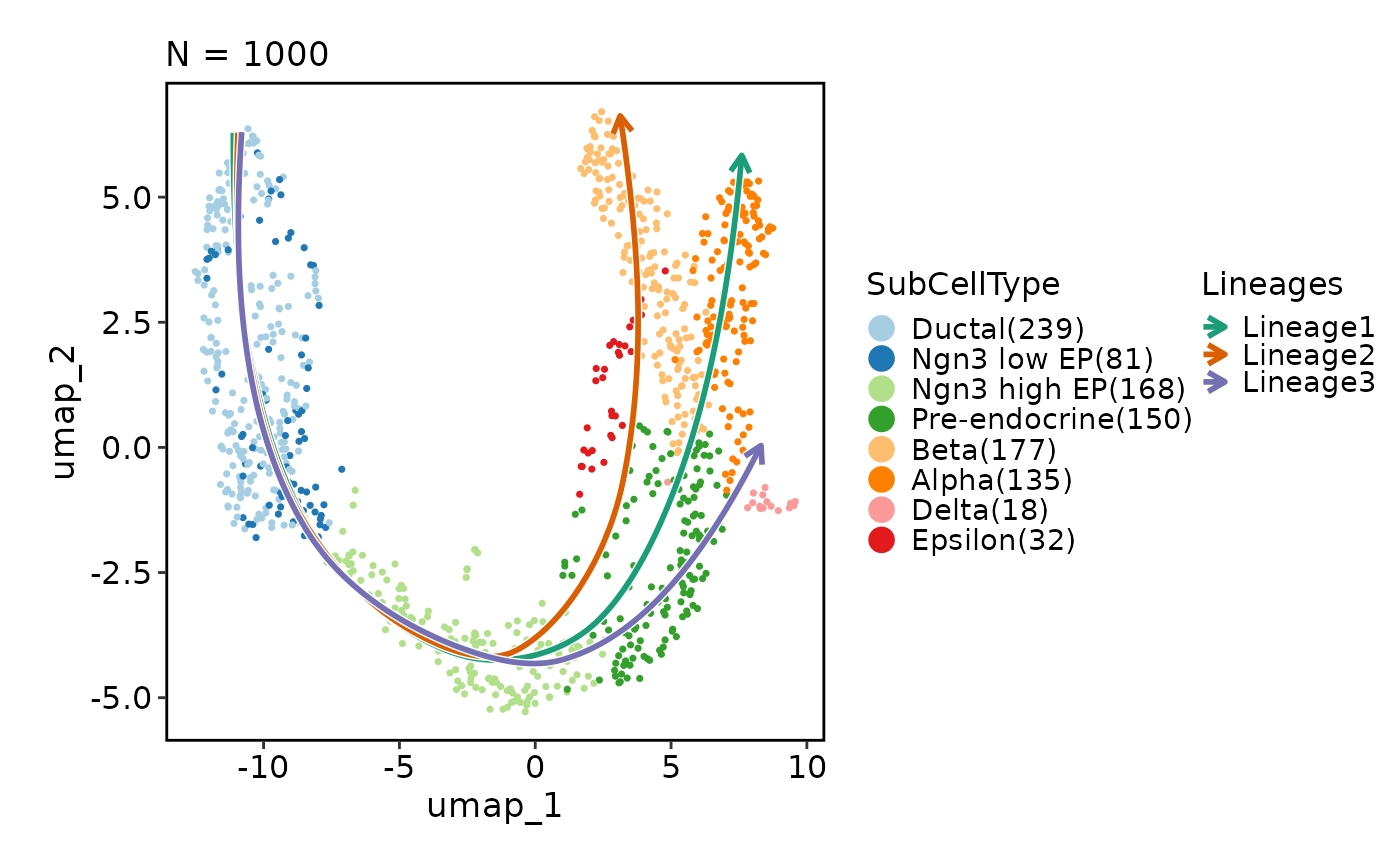
 # Show lineages on the plot based on the pseudotime
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
lineages = paste0("Lineage", 1:3))
#> Warning: Removed 8 rows containing missing values or values outside the scale range
#> (`geom_path()`).
#> Warning: Removed 8 rows containing missing values or values outside the scale range
#> (`geom_path()`).
# Show lineages on the plot based on the pseudotime
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
lineages = paste0("Lineage", 1:3))
#> Warning: Removed 8 rows containing missing values or values outside the scale range
#> (`geom_path()`).
#> Warning: Removed 8 rows containing missing values or values outside the scale range
#> (`geom_path()`).
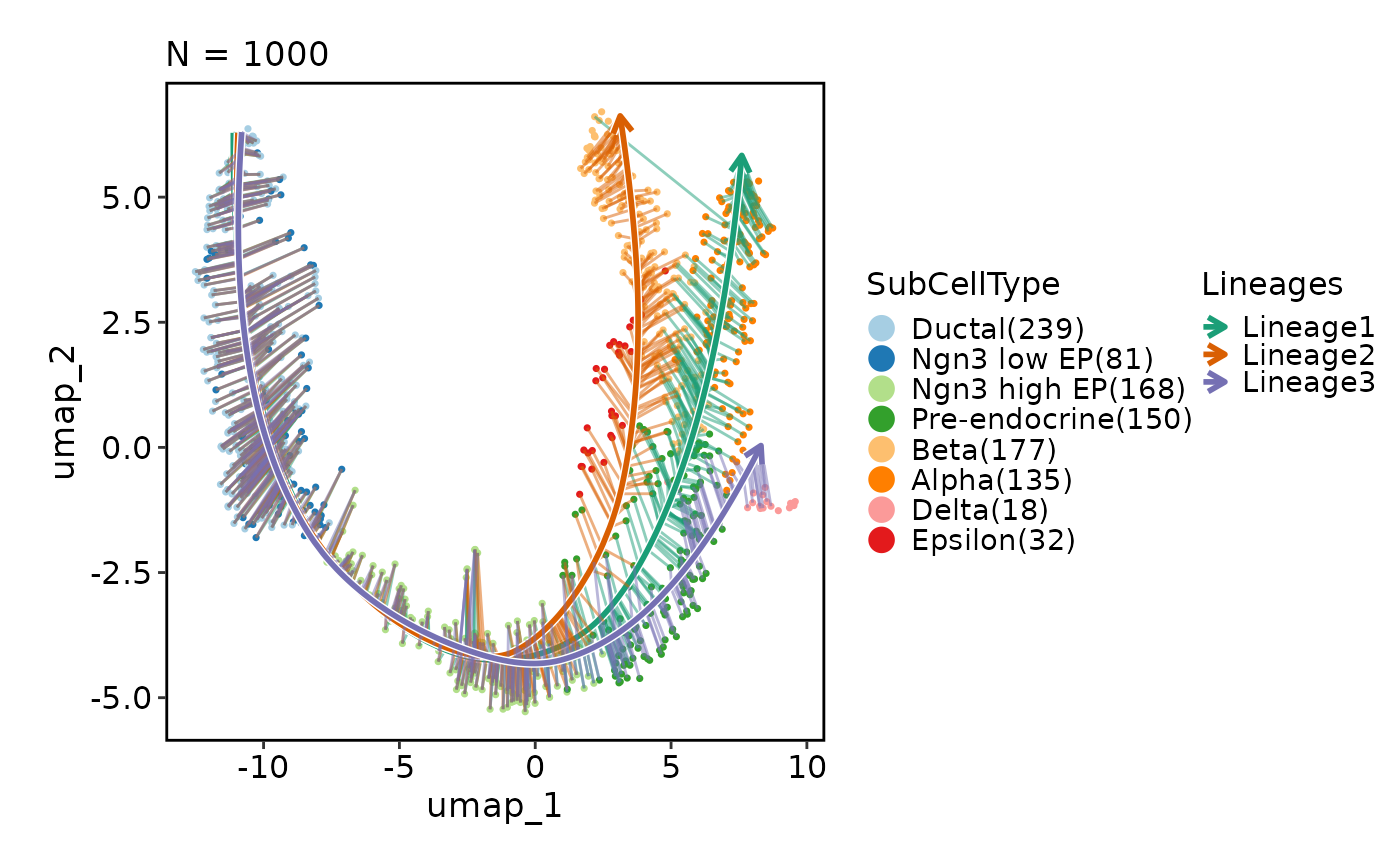
 CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
lineages = paste0("Lineage", 1:3), lineages_whiskers = TRUE)
#> Warning: Removed 8 rows containing missing values or values outside the scale range
#> (`geom_segment()`).
#> Warning: Removed 8 rows containing missing values or values outside the scale range
#> (`geom_path()`).
#> Warning: Removed 8 rows containing missing values or values outside the scale range
#> (`geom_path()`).
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
lineages = paste0("Lineage", 1:3), lineages_whiskers = TRUE)
#> Warning: Removed 8 rows containing missing values or values outside the scale range
#> (`geom_segment()`).
#> Warning: Removed 8 rows containing missing values or values outside the scale range
#> (`geom_path()`).
#> Warning: Removed 8 rows containing missing values or values outside the scale range
#> (`geom_path()`).
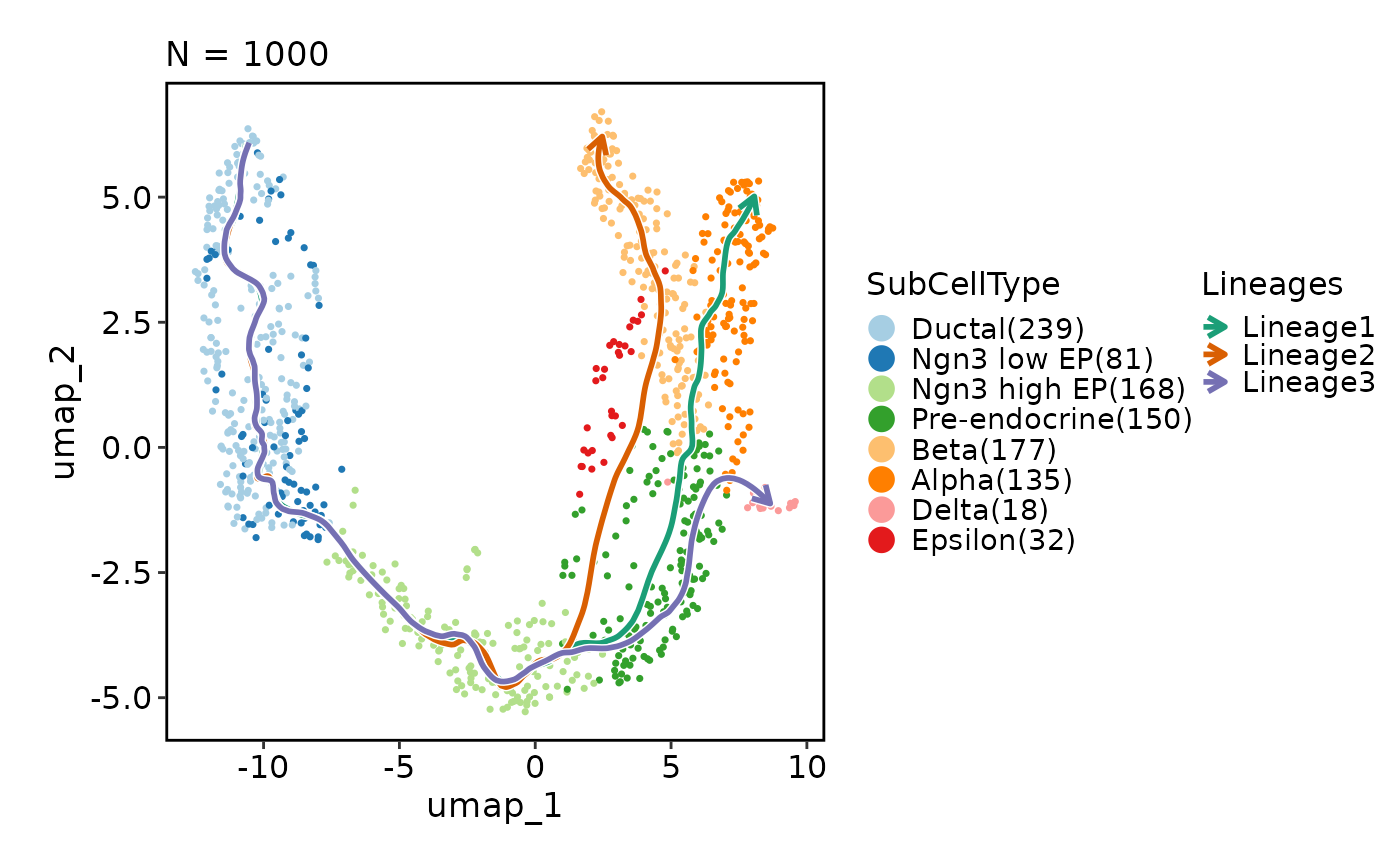
 CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
lineages = paste0("Lineage", 1:3), lineages_span = 0.1)
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "UMAP",
lineages = paste0("Lineage", 1:3), lineages_span = 0.1)
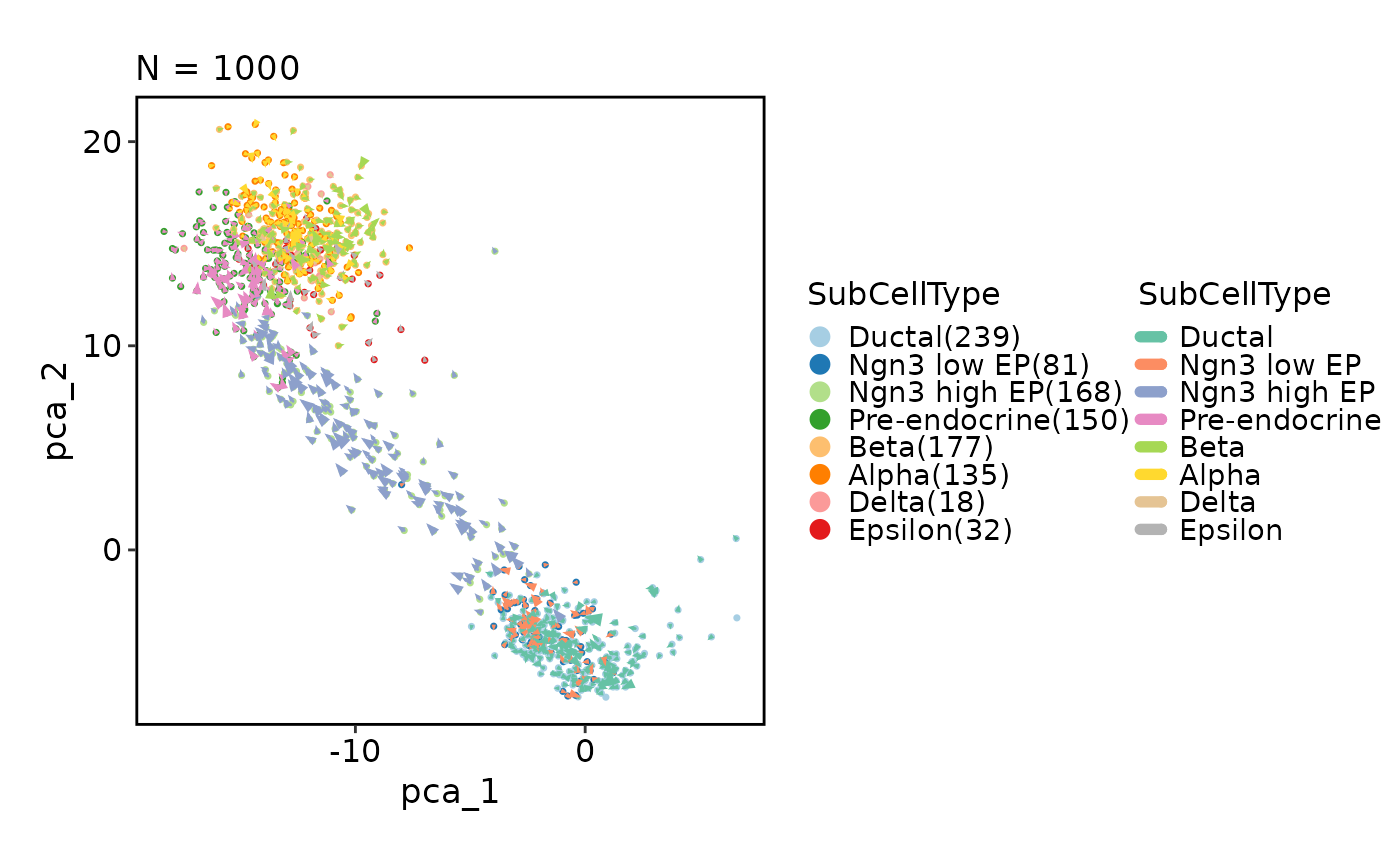
 # Velocity
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "PCA",
velocity = "stochastic_PCA")
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_segment()`).
# Velocity
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "PCA",
velocity = "stochastic_PCA")
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_segment()`).
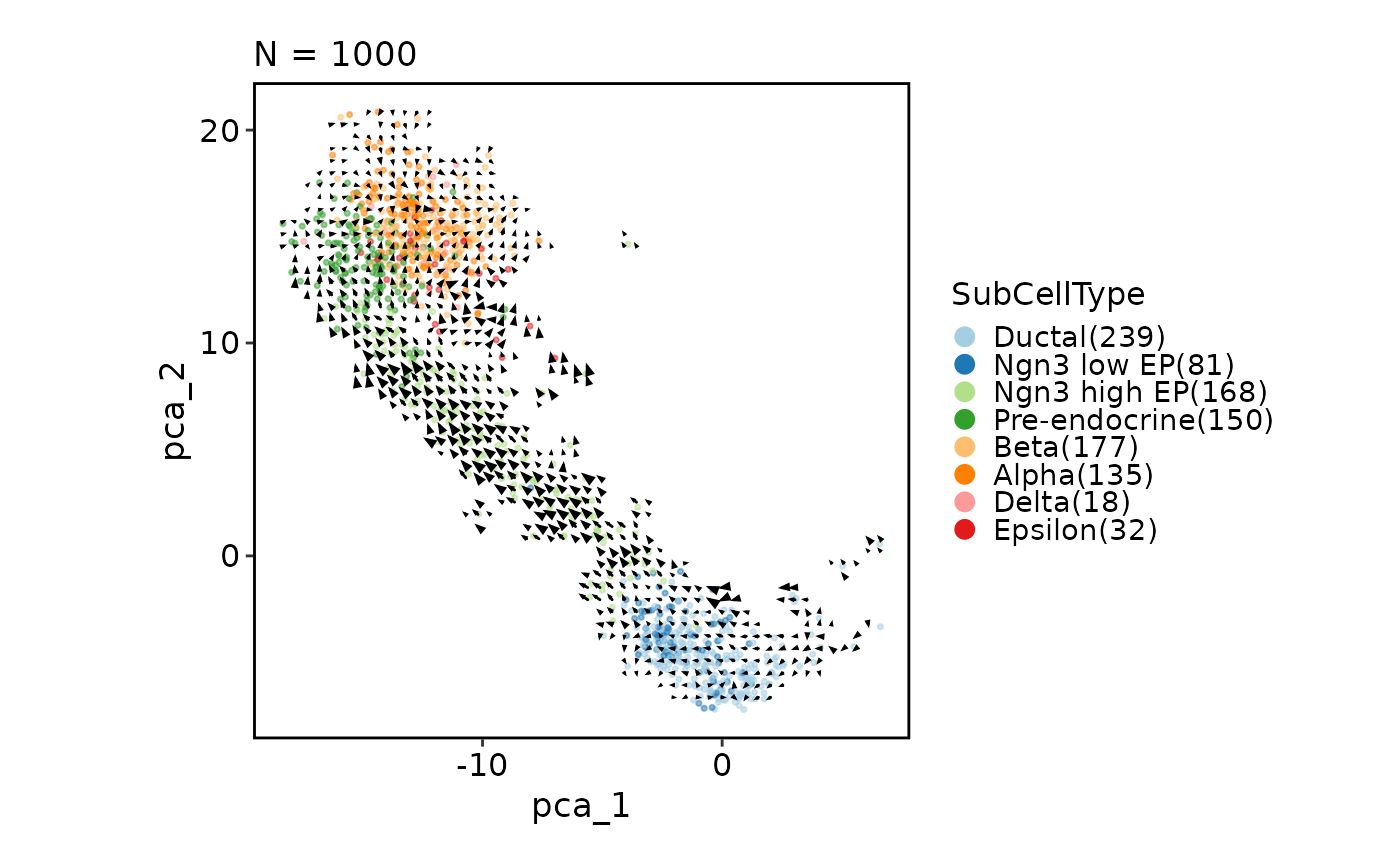
 CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "PCA",
velocity = "stochastic_PCA", velocity_plot_type = "grid", pt_alpha = 0.5)
#> Warning: Removed 15 rows containing missing values or values outside the scale range
#> (`geom_segment()`).
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "PCA",
velocity = "stochastic_PCA", velocity_plot_type = "grid", pt_alpha = 0.5)
#> Warning: Removed 15 rows containing missing values or values outside the scale range
#> (`geom_segment()`).
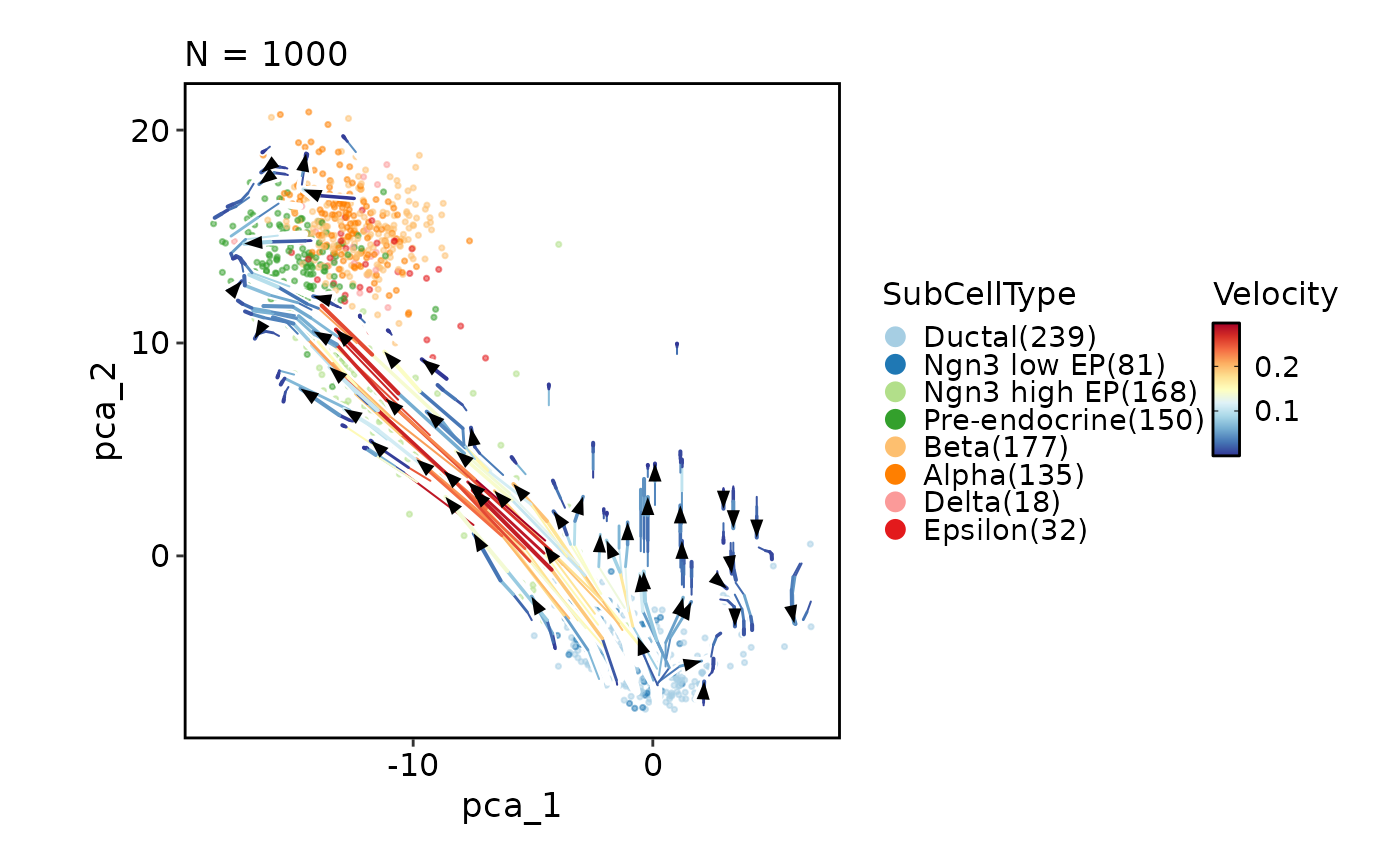
 CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "PCA",
velocity = "stochastic_PCA", velocity_plot_type = "stream", pt_alpha = 0.5)
#> Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` instead.
#> ℹ The deprecated feature was likely used in the plotthis package.
#> Please report the issue at <https://github.com/pwwang/plotthis/issues>.
CellDimPlot(pancreas_sub, group_by = "SubCellType", reduction = "PCA",
velocity = "stochastic_PCA", velocity_plot_type = "stream", pt_alpha = 0.5)
#> Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` instead.
#> ℹ The deprecated feature was likely used in the plotthis package.
#> Please report the issue at <https://github.com/pwwang/plotthis/issues>.
 # }
# }