An R package for enrichment (over-representation) analysis of gene sets. It reproduces the results by clusterProfiler and enrichr, but with following features:
- Offline (no internet connection required)
- Analysis against custom gene sets (GMT files)
- Both styles (clusterProfiler and enrichr) supported
- Seemless integration with
scplotterfor visualization
Installation
You can install the development version of enrichit from GitHub with:
# install.packages("devtools")
devtools::install_github("pwwang/enrichit")
# or remotes::install_github("pwwang/enrichit")Usage
library(enrichit)
data(userlist)
kegg_gmt <- system.file("extdata", "KEGG_2021_Human.gmt.gz", package = "enrichit")
hallmark_gmt <- system.file("extdata", "MSigDB_Hallmark_2020.gmt.gz", package = "enrichit")
EnrichIt(userlist, c(kegg_gmt, hallmark_gmt))
# Term Overlap P.value
# 202 Pancreatic secretion 29/102 1.462598e-13
# 161 Maturity onset diabetes of the young 14/26 1.403986e-11
# 230 Protein digestion and absorption 20/103 9.254422e-07
# 178 Neuroactive ligand-receptor interaction 41/341 3.071673e-06
# 45 Carbohydrate digestion and absorption 11/47 4.410201e-05
# 87 Fat digestion and absorption 10/43 1.037564e-04
# 501 Pancreas Beta Cells 19/40 5.821496e-14
# 49 KRAS Signaling Dn 20/200 8.805898e-03
# 451 Pperoxisome 7/104 3.681308e-01
# 441 Bile Acid Metabolism 7/112 4.427207e-01
# 22 Hedgehog Signaling 2/36 6.087590e-01
# 401 Angiogenesis 2/36 6.087590e-01
# ... ...
# Adjusted.P.value Odds.Ratio Combined.Score
# 202 3.510235e-11 6.9503571 205.4066273
# 161 1.684783e-09 20.0200000 500.2821950
# 230 7.403538e-05 4.1479048 57.6268164
# 178 1.843004e-04 2.3870157 30.2990782
# 45 2.116896e-03 5.2050857 52.2018318
# 87 4.150256e-03 5.1550271 47.2894589
# 501 1.921094e-12 15.6424970 476.6993715
# 49 1.452973e-01 1.8945006 8.9654086
# 451 9.999916e-01 1.2139642 1.2131352
# 441 9.999916e-01 1.1205931 0.9130774
# 22 9.999916e-01 0.9875048 0.4901311
# 401 9.999916e-01 0.9875048 0.4901311
# ... ...
# Genes Rank Database
# 202 AMY1B;PRSS2;SLC4A4;... 1 KEGG_2021_Human
# 161 PDX1;BHLHA15;NEUROD1;... 2 KEGG_2021_Human
# 230 PRSS2;PRSS1;SLC8A2;... 3 KEGG_2021_Human
# 178 SST;AGTR2;CNR1;... 4 KEGG_2021_Human
# 45 AMY1B;G6PC1;SI;... 5 KEGG_2021_Human
# 87 PLA2G2D;PLA2G2A;PLA2G4A;... 6 KEGG_2021_Human
# 501 CHGA;ABCC8;NEUROD1;... 1 MSigDB_Hallmark_2020
# 49 EGF;CNTFR;NOS1;... 2 MSigDB_Hallmark_2020
# 451 RXRG;CEL;CACNA1B;ABCC8;ALB;SERPINA6;TTR 3 MSigDB_Hallmark_2020
# 441 RXRG;NR0B2;SERPINA6;NR1H4;GNMT;TTR;GC 4 MSigDB_Hallmark_2020
# 22 CNTFR;NKX6-1 5 MSigDB_Hallmark_2020
# 401 VTN;APOH 5 MSigDB_Hallmark_2020
# ... ...
EnrichIt(userlist, c(kegg_gmt, hallmark_gmt), style = "clusterProfiler")
# ID Description GeneRatio
# 202 KEGG_2021_Human_202 Pancreatic secretion 29/279
# 178 KEGG_2021_Human_178 Neuroactive ligand-receptor interaction 41/279
# 161 KEGG_2021_Human_161 Maturity onset diabetes of the young 14/279
# 230 KEGG_2021_Human_230 Protein digestion and absorption 20/279
# 45 KEGG_2021_Human_45 Carbohydrate digestion and absorption 11/279
# 87 KEGG_2021_Human_87 Fat digestion and absorption 10/279
# 501 MSigDB_Hallmark_2020_50 Pancreas Beta Cells 19/120
# 49 MSigDB_Hallmark_2020_49 KRAS Signaling Dn 20/120
# 321 MSigDB_Hallmark_2020_32 Xenobiotic Metabolism 10/120
# 451 MSigDB_Hallmark_2020_45 Pperoxisome 7/120
# 441 MSigDB_Hallmark_2020_44 Bile Acid Metabolism 7/120
# 231 MSigDB_Hallmark_2020_23 Complement 9/120
# BgRatio pvalue p.adjust qvalue geneID
# 202 102/10922 7.021146e-23 1.685075e-20 1.345104e-20 AMY1B/PRSS2/PRSS1/...
# 178 341/10922 5.947771e-17 7.137325e-15 5.697338e-15 SST/AGTR2/PRSS2/...
# 161 26/10922 2.674313e-16 2.139450e-14 1.707807e-14 PDX1/BHLHA15/HNF1B/...
# 230 103/10922 1.091957e-12 6.551741e-11 5.229898e-11 PRSS2/PRSS1/ATP1A2/...
# 45 47/10922 1.910985e-08 9.172727e-07 7.322089e-07 AMY1B/G6PC1/G6PC2/...
# 87 43/10922 9.170990e-08 3.668396e-06 2.928281e-06 PLA2G2D/PLA2G2A/CEL/...
# 501 40/10922 1.471191e-27 4.854929e-26 3.097243e-26 CHGA/ABCC8/PDX1/...
# 49 200/10922 4.117838e-14 6.794432e-13 4.334566e-13 EGF/CNTFR/TENT5C/...
# 321 200/10922 6.837635e-05 7.521398e-04 4.798340e-04 RBP4/ITIH4/PDK4/...
# 451 104/10922 1.428239e-04 1.178297e-03 7.517049e-04 RXRG/CEL/CACNA1B/...
# 441 112/10922 2.265978e-04 1.495545e-03 9.540959e-04 RXRG/NR0B2/SERPINA6/...
# 231 200/10922 3.505544e-04 1.928049e-03 1.230015e-03 PRSS3/KLKB1/KLK1/...
# Count Database
# 202 29 KEGG_2021_Human
# 178 41 KEGG_2021_Human
# 161 14 KEGG_2021_Human
# 230 20 KEGG_2021_Human
# 45 11 KEGG_2021_Human
# 87 10 KEGG_2021_Human
# 501 19 MSigDB_Hallmark_2020
# 49 20 MSigDB_Hallmark_2020
# 321 10 MSigDB_Hallmark_2020
# 451 7 MSigDB_Hallmark_2020
# 441 7 MSigDB_Hallmark_2020
# 231 9 MSigDB_Hallmark_2020Visualization
library(scplotter)
library(enrichit)
data(userlist)
kegg_gmt <- system.file("extdata", "KEGG_2021_Human.gmt.gz", package = "enrichit")
hallmark_gmt <- system.file("extdata", "MSigDB_Hallmark_2020.gmt.gz", package = "enrichit")
enrich_result <- EnrichIt(userlist, c(kegg_gmt, hallmark_gmt), style = "clusterProfiler")
EnrichmentPlot(enrich_result, split_by = "Database")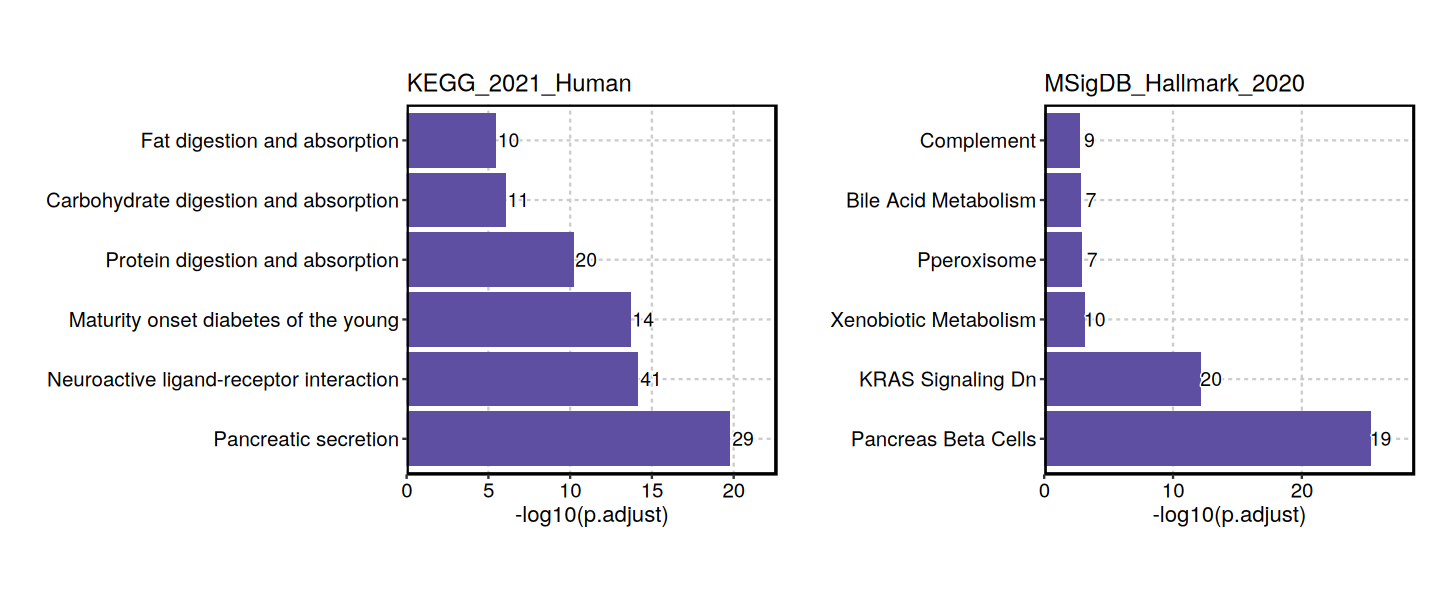
EnrichmentPlot(enrich_result, plot_type = "dot", split_by = "Database")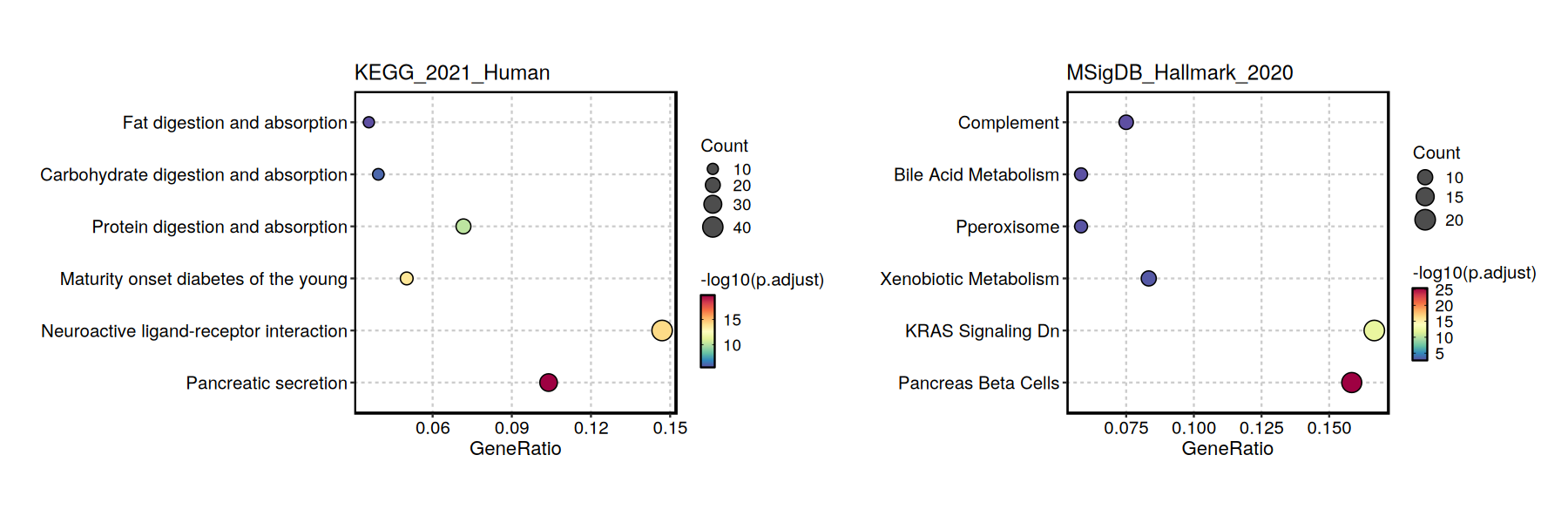
EnrichmentPlot(enrich_result[enrich_result$Database == "KEGG_2021_Human", ], plot_type = "network")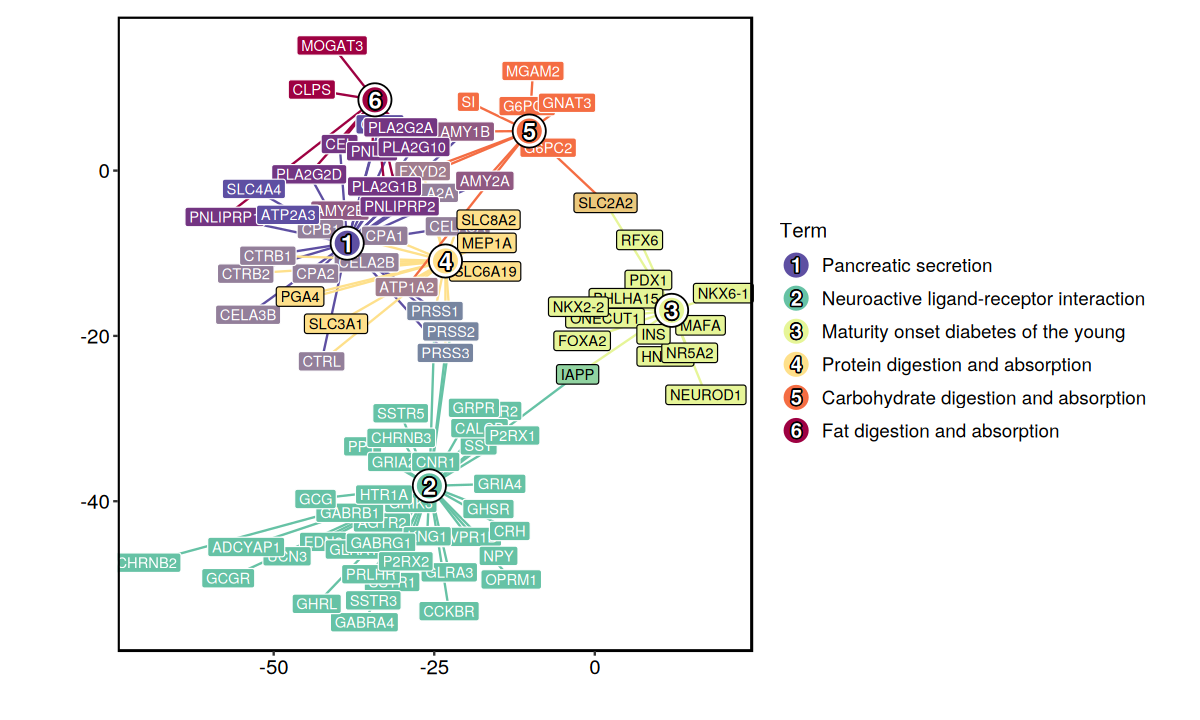
EnrichmentPlot(enrich_result, plot_type = "wordcloud")
Documentation
See documentation for more details.