Over-representation Enrichment Analysis
Source:vignettes/over-representation_enrichment_analysis.Rmd
over-representation_enrichment_analysis.RmdIntroduction
This vignette demonstrates how to perform over-representation
enrichment analysis using the enrichit package. The package
provides a convenient way to analyze gene sets against custom gene sets
(GMT files) and supports both clusterProfiler and
enrichr styles of analysis.
The package is designed to work offline, making it suitable for
environments with limited internet access. Additionally, it seamlessly
integrates with the scplotter
package for visualization.
Installation
You can install the development version of enrichit from
GitHub with:
# install.packages("devtools")
devtools::install_github("pwwang/enrichit")
# remotes::install_github("pwwang/enrichit")Data preparation
Load example data
The example data used in this vignette is included in the package. You can load it using the following command:
data(userlist)For this example, we will use the userlist data, which
contains a list of genes of interest. The data is a character vector of
gene symbols. You can replace this with your own gene list. They don’t
have to be gene symbols, but they should match the gene names in the GMT
files you are using for analysis.
Load GMT files
The enrichit package comes with built-in GMT files for
BioCarta, GO, KEGG, MSigDB Hallmark, Reactome pathways and WikiPathways.
You can find these files in the extdata directory of the
package. You can find more public GMT files in the MSigDB database or
the enrichr
database. You can also use your own custom GMT files.
To load the GMT files, you can use the following commands:
kegg_gmt <- system.file("extdata", "KEGG_2021_Human.gmt.gz", package = "enrichit")
hallmark_gmt <- system.file("extdata", "MSigDB_Hallmark_2020.gmt.gz", package = "enrichit")
pathways <- ParseGMT(kegg_gmt)
pathways[1:2]
#> $`ABC transporters`
#> [1] "ABCA2" "ABCC4" "ABCG8" "ABCA3" "ABCC5" "ABCC2" "ABCA1" "ABCC3"
#> [9] "ABCA6" "ABCC8" "ABCA7" "ABCC9" "ABCA4" "ABCC6" "ABCA5" "TAP2"
#> [17] "ABCA8" "TAP1" "ABCA9" "ABCA10" "ABCB10" "ABCA12" "ABCB11" "ABCC10"
#> [25] "ABCG1" "ABCG4" "ABCC1" "ABCG5" "ABCG2" "CFTR" "ABCB4" "ABCB1"
#> [33] "ABCD3" "ABCD4" "ABCB7" "ABCB8" "ABCB5" "ABCB6" "ABCB9" "ABCA13"
#> [41] "ABCC11" "ABCC12" "DEFB1" "ABCD1" "ABCD2"
#>
#> $`AGE-RAGE signaling pathway in diabetic complications`
#> [1] "TGFB1" "TGFB3" "TGFB2" "VCAM1" "AGT" "MAPK13"
#> [7] "MAPK14" "COL1A2" "MAPK11" "COL1A1" "MAPK12" "MAPK10"
#> [13] "AGTR1" "BAX" "PLCD1" "PLCD4" "PLCD3" "RELA"
#> [19] "SERPINE1" "TNF" "FOXO1" "NRAS" "CCND1" "AKT3"
#> [25] "AKT1" "PLCE1" "AKT2" "EGR1" "SMAD3" "STAT5B"
#> [31] "SMAD2" "SMAD4" "PRKCD" "PRKCE" "CYBB" "PRKCB"
#> [37] "PRKCA" "SELE" "NFKB1" "DIAPH1" "PRKCZ" "PIK3R1"
#> [43] "NOX4" "NOX1" "AGER" "ICAM1" "PIM1" "PLCG1"
#> [49] "PLCG2" "STAT5A" "EDN1" "JUN" "MMP2" "VEGFC"
#> [55] "FN1" "VEGFD" "NFATC1" "TGFBR1" "TGFBR2" "VEGFA"
#> [61] "VEGFB" "IL1A" "IL1B" "COL3A1" "IL6" "PIK3CB"
#> [67] "PIK3CA" "CDK4" "BCL2" "CXCL8" "PIK3R3" "PIK3R2"
#> [73] "CASP3" "NOS3" "STAT1" "STAT3" "F3" "CDC42"
#> [79] "COL4A1" "PLCB4" "COL4A3" "COL4A2" "COL4A5" "COL4A4"
#> [85] "COL4A6" "KRAS" "PLCB2" "PLCB3" "PLCB1" "CDKN1B"
#> [91] "PIK3CD" "MAPK9" "THBD" "MAPK8" "MAPK3" "CCL2"
#> [97] "HRAS" "MAPK1" "RAC1" "JAK2"Specifying a URL of a GMT file is also supported. For example, you can use the following command to load a GMT file from a URL:
# gmt_url <- "https://example.com/path/to/your/gmt/file.gmt"
# pathways <- ParseGMT(gmt_url)Additionally, you can also use a name of a library from the enrichr
database to load the GMT file. If they are one of the built-in libraries
(BioCarta, BioCarta_2016,
GO_Biological_Process,
GO_Biological_Process_2025,
GO_Cellular_Component,
GO_Cellular_Component_2025,
GO_Molecular_Function,
GO_Molecular_Function_2025, KEGG,
KEGG_2021, KEGG_Human,
KEGG_2021_Human, Hallmark,
MSigDB_Hallmark, MSigDB_Hallmark_2020,
Reactome, Reactome_Pathways,
Reactome_Pathways_2024, WikiPathways,
WikiPathways_2024), the GMT file will be loaded from the
package. Otherwise, it will be downloaded from the enrichr
database library.
# Load built-in libraries
library_name <- "MSigDB_Hallmark"
hallmarks <- ParseGMT(library_name)
hallmarks[1:2]
#> $`TNF-alpha Signaling via NF-kB`
#> [1] "MARCKS" "IL23A" "NINJ1" "TNFSF9" "SIK1" "ATF3"
#> [7] "SERPINE1" "MYC" "HES1" "CCN1" "CCNL1" "EGR1"
#> [13] "EGR2" "EGR3" "JAG1" "ABCA1" "GADD45B" "GADD45A"
#> [19] "KLF10" "PLK2" "EIF1" "EHD1" "FOSL2" "FOSL1"
#> [25] "GPR183" "PLPP3" "IFIT2" "ICAM1" "ZC3H12A" "IER2"
#> [31] "IL12B" "IER5" "JUNB" "IER3" "STAT5A" "DUSP5"
#> [37] "EDN1" "DUSP4" "JUN" "DUSP1" "DUSP2" "TSC22D1"
#> [43] "CCL20" "SPHK1" "LIF" "IL18" "TUBB2A" "RHOB"
#> [49] "VEGFA" "IL1A" "PTPRE" "TLR2" "IL1B" "BHLHE40"
#> [55] "CLCF1" "ID2" "REL" "FJX1" "SGK1" "BTG3"
#> [61] "BTG2" "BTG1" "SDC4" "LITAF" "AREG" "SOCS3"
#> [67] "PANX1" "RIPK2" "NFIL3" "SERPINB2" "GCH1" "IFNGR2"
#> [73] "G0S2" "FOS" "F3" "SERPINB8" "SPSB1" "FOSB"
#> [79] "PER1" "F2RL1" "HBEGF" "CD44" "TRIP10" "CDKN1A"
#> [85] "PTGER4" "PTGS2" "IFIH1" "NAMPT" "OLR1" "ICOSLG"
#> [91] "PHLDA1" "ZBTB10" "TAP1" "PNRC1" "CXCL10" "CXCL11"
#> [97] "IL6ST" "CD69" "SQSTM1" "RELA" "CD83" "CSF2"
#> [103] "CSF1" "CD80" "PPP1R15A" "TNC" "TNF" "RELB"
#> [109] "TANK" "ZFP36" "CCND1" "RNF19B" "CCRL2" "DENND5A"
#> [115] "MAP3K8" "PHLDA2" "LDLR" "SLC16A6" "SMAD3" "TGIF1"
#> [121] "MAP2K3" "DDX58" "INHBA" "TRAF1" "NFKB1" "NFKB2"
#> [127] "GEM" "MAFF" "NR4A3" "RCAN1" "EFNA1" "NR4A2"
#> [133] "MXD1" "BIRC2" "BIRC3" "YRDC" "IL7R" "PFKFB3"
#> [139] "IRS2" "SLC2A3" "PLAU" "SAT1" "SLC2A6" "ETS2"
#> [145] "NR4A1" "SNN" "PMEPA1" "TNFRSF9" "MSC" "TIPARP"
#> [151] "LAMB3" "GFPT2" "CFLAR" "TNIP1" "IRF1" "NFKBIA"
#> [157] "BMP2" "IL6" "TNIP2" "BCL6" "BCL3" "NFKBIE"
#> [163] "NFE2L2" "B4GALT1" "NFAT5" "BCL2A1" "TNFAIP8" "TNFAIP6"
#> [169] "TNFAIP3" "CXCL2" "CXCL1" "TNFAIP2" "CXCL3" "CXCL6"
#> [175] "FUT4" "DRAM1" "DNAJB4" "PDE4B" "PDLIM5" "MCL1"
#> [181] "KDM6B" "IL15RA" "PLAUR" "ATP2B1" "KLF4" "KLF2"
#> [187] "SOD2" "KLF9" "KLF6" "ACKR3" "PTX3" "B4GALT5"
#> [193] "TRIB1" "CEBPB" "CEBPD" "PLEK" "CCL5" "KYNU"
#> [199] "CCL4" "CCL2"
#>
#> $Hypoxia
#> [1] "WSB1" "JMJD6" "XPNPEP1" "RRAGD" "PPP1R3C" "MT1E"
#> [7] "TES" "ATF3" "SERPINE1" "GLRX" "UGP2" "NOCT"
#> [13] "MXI1" "DPYSL4" "CCN5" "CCN2" "PPARGC1A" "CCN1"
#> [19] "DTNA" "SIAH2" "MIF" "PRKCA" "EXT1" "FOSL2"
#> [25] "SLC6A6" "SULT2B1" "BCAN" "LALBA" "CHST2" "SDC2"
#> [31] "HOXB9" "CHST3" "LXN" "NEDD4L" "PDGFB" "ENO1"
#> [37] "ENO2" "ENO3" "RBPJ" "MT2A" "PLAC8" "PRDX5"
#> [43] "HMOX1" "PDK1" "IER3" "PDK3" "EDN2" "ANXA2"
#> [49] "JUN" "DUSP1" "TPI1" "AMPD3" "CP" "VEGFA"
#> [55] "P4HA1" "P4HA2" "BHLHE40" "CCNG2" "PAM" "ALDOC"
#> [61] "ALDOB" "ALDOA" "GPI" "BTG1" "CITED2" "SDC3"
#> [67] "SDC4" "STC1" "ADM" "STC2" "VLDLR" "BRS3"
#> [73] "LDHC" "LDHA" "CSRP2" "IDS" "NFIL3" "PCK1"
#> [79] "TPD52" "PKLR" "PGAM2" "GAA" "FOS" "BGN"
#> [85] "F3" "TPST2" "ANGPTL4" "PLIN2" "INHA" "ERRFI1"
#> [91] "SRPX" "CDKN1B" "CDKN1C" "ERO1A" "HEXA" "CDKN1A"
#> [97] "NCAN" "GBE1" "GCNT2" "PYGM" "GYS1" "NDST2"
#> [103] "NDST1" "AKAP12" "STBD1" "CXCR4" "CA12" "HS3ST1"
#> [109] "MAP3K1" "TGFB3" "CAVIN1" "CAVIN3" "KLHL24" "PNRC1"
#> [115] "SAP30" "GCK" "ILVBL" "COL5A1" "TGFBI" "GAPDHS"
#> [121] "S100A4" "PKP1" "PPP1R15A" "HDLBP" "EGFR" "FOXO3"
#> [127] "NDRG1" "KIF5A" "ZFP36" "GPC1" "HAS1" "GPC4"
#> [133] "GPC3" "PPFIA4" "IGFBP1" "BNIP3L" "IGFBP3" "B3GALT6"
#> [139] "PGF" "MAFF" "EFNA1" "ISG20" "EFNA3" "LOX"
#> [145] "PFKFB3" "IRS2" "SLC2A1" "AK4" "SLC2A3" "SLC2A5"
#> [151] "ETS1" "RORA" "TGM2" "FAM162A" "PIM1" "ANKZF1"
#> [157] "TIPARP" "HSPA5" "CAV1" "GALK1" "GRHPR" "IL6"
#> [163] "ADORA2B" "MYH9" "BCL2" "FBP1" "TMEM45A" "SLC25A1"
#> [169] "TNFAIP3" "HK1" "HK2" "CASP6" "PGK1" "B4GALNT2"
#> [175] "SLC37A4" "TKTL1" "TPBG" "PLAUR" "DCN" "SELENBP1"
#> [181] "KLF7" "NAGK" "KLF6" "PFKL" "DDIT4" "DDIT3"
#> [187] "ACKR3" "KDELR3" "GAPDH" "PFKP" "KDM3A" "ZNF292"
#> [193] "NR3C1" "PHKG1" "VHL" "ATP7A" "PGM2" "SCARB1"
#> [199] "LARGE1" "PGM1"
# Load from enrichr library using name
library_name <- "ChEA_2022"
chea <- ParseGMT(library_name)
#> Using cached file: /home/runner/.cache/R/enrichit/3d40e13b/ChEA_2022.gmt
chea[1:2]
#> $`BP1 19119308 ChIP-ChIP Hs578T Human`
#> [1] "CBR1" "OR1A1" "HTR1B" "LAPTM5" "ADAM21" "HPSE2" "SELE"
#> [8] "CSF2RB" "VEGFA" "PTPRE" "LSM7" "IL2RA" "MMP27" "LGALS13"
#> [15] "CCL4" "ITGA9" "CASQ1" "TAC3"
#>
#> $`FOXP1 22492998 ChIP-Seq STRATIUM Mouse`
#> [1] "MCTS2" "PCSK2" "BCL11B" "ALMS1" "ETFDH" "PTEN" "RLF"
#> [8] "ACYP2" "RXFP1" "FOXP1" "RPL36AL" "TMEM125" "TMX4" "MGAT2"
#> [15] "IGFBP7" "LDLR"Enrichment analysis
Single gene set database
# pathways was loaded previously
# pathways = ParseGMT(kegg_gmt)
head(EnrichIt(userlist, pathways), n = 3)
#> Warning in CheckUserList(userlist, db, use_matched_only = use_matched_only):
#> User list contains 335 unmatched genes. Showing first 10: A1CF, A4GNT, AADAC,
#> ADAMTS8, ADGRV1, AGR3, AJAP1, AKAIN1, ALKAL2, AMBP
#> Term Overlap P.value
#> 202 Pancreatic secretion 29/102 2.084613e-20
#> 161 Maturity onset diabetes of the young 14/26 3.925575e-15
#> 178 Neuroactive ligand-receptor interaction 41/341 8.073882e-14
#> Adjusted.P.value Odds.Ratio Combined.Score
#> 202 5.003072e-18 13.115022 594.3350
#> 161 4.710689e-13 37.671667 1249.6168
#> 178 6.459105e-12 4.552216 137.2382
#> Genes
#> 202 AMY1B;PRSS2;PRSS1;ATP1A2;PRSS3;CELA2A;CELA2B;CPA2;CPA1;CEL;CFTR;PNLIP;CELA3A;CELA3B;AMY2A;AMY2B;CPB1;PLA2G2D;PLA2G2A;CTRL;FXYD2;PNLIPRP2;PNLIPRP1;CTRB2;PLA2G10;CTRB1;PLA2G1B;ATP2A3;SLC4A4
#> 161 PDX1;BHLHA15;HNF1B;MAFA;NR5A2;IAPP;RFX6;FOXA2;ONECUT1;SLC2A2;INS;NKX2-2;NKX6-1;NEUROD1
#> 178 SST;AGTR2;PRSS2;VIPR2;PRSS1;PRSS3;AVPR1B;CCKBR;GRPR;PPY;EDN3;GABRA4;GABRB1;GRIK3;GCGR;GHRL;GLRA1;GLRA3;UCN3;NPY;CHRNB3;CHRNB2;GCG;IAPP;GHSR;GRIA2;CALCB;GRIA4;OPRM1;SSTR1;HTR1A;SSTR3;SSTR5;KNG1;PRLHR;GABRG1;ADCYAP1;P2RX2;P2RX1;CRH;CNR1
#> Rank Database
#> 202 1 pathways
#> 161 2 pathways
#> 178 3 pathwaysYou can also specify the GMT file path directly:
# kegg_gmt <- system.file("extdata", "KEGG_2021_Human.gmt.gz", package = "enrichit")
head(EnrichIt(userlist, kegg_gmt), n = 3)
#> Warning in CheckUserList(userlist, db, use_matched_only = use_matched_only):
#> User list contains 335 unmatched genes. Showing first 10: A1CF, A4GNT, AADAC,
#> ADAMTS8, ADGRV1, AGR3, AJAP1, AKAIN1, ALKAL2, AMBP
#> Term Overlap P.value
#> 202 Pancreatic secretion 29/102 2.084613e-20
#> 161 Maturity onset diabetes of the young 14/26 3.925575e-15
#> 178 Neuroactive ligand-receptor interaction 41/341 8.073882e-14
#> Adjusted.P.value Odds.Ratio Combined.Score
#> 202 5.003072e-18 13.115022 594.3350
#> 161 4.710689e-13 37.671667 1249.6168
#> 178 6.459105e-12 4.552216 137.2382
#> Genes
#> 202 AMY1B;PRSS2;PRSS1;ATP1A2;PRSS3;CELA2A;CELA2B;CPA2;CPA1;CEL;CFTR;PNLIP;CELA3A;CELA3B;AMY2A;AMY2B;CPB1;PLA2G2D;PLA2G2A;CTRL;FXYD2;PNLIPRP2;PNLIPRP1;CTRB2;PLA2G10;CTRB1;PLA2G1B;ATP2A3;SLC4A4
#> 161 PDX1;BHLHA15;HNF1B;MAFA;NR5A2;IAPP;RFX6;FOXA2;ONECUT1;SLC2A2;INS;NKX2-2;NKX6-1;NEUROD1
#> 178 SST;AGTR2;PRSS2;VIPR2;PRSS1;PRSS3;AVPR1B;CCKBR;GRPR;PPY;EDN3;GABRA4;GABRB1;GRIK3;GCGR;GHRL;GLRA1;GLRA3;UCN3;NPY;CHRNB3;CHRNB2;GCG;IAPP;GHSR;GRIA2;CALCB;GRIA4;OPRM1;SSTR1;HTR1A;SSTR3;SSTR5;KNG1;PRLHR;GABRG1;ADCYAP1;P2RX2;P2RX1;CRH;CNR1
#> Rank Database
#> 202 1 KEGG_2021_Human
#> 161 2 KEGG_2021_Human
#> 178 3 KEGG_2021_HumanYou may notice that the values of the Database column is
different. We keep that column to support enrichment analysis against
multiple databases. When no database is specified (the first
EnrichIt example), the argument name
(pathways) is used as the database name. When a GMT file is
specified (the second EnrichIt example), the GMT file name
is used as the database name. You can also explictly specify the
database name:
head(EnrichIt(userlist, list(KEGG = pathways)), n = 3)
#> Warning in CheckUserList(userlist, db, use_matched_only = use_matched_only):
#> User list contains 335 unmatched genes. Showing first 10: A1CF, A4GNT, AADAC,
#> ADAMTS8, ADGRV1, AGR3, AJAP1, AKAIN1, ALKAL2, AMBP
#> Term Overlap P.value
#> 202 Pancreatic secretion 29/102 2.084613e-20
#> 161 Maturity onset diabetes of the young 14/26 3.925575e-15
#> 178 Neuroactive ligand-receptor interaction 41/341 8.073882e-14
#> Adjusted.P.value Odds.Ratio Combined.Score
#> 202 5.003072e-18 13.115022 594.3350
#> 161 4.710689e-13 37.671667 1249.6168
#> 178 6.459105e-12 4.552216 137.2382
#> Genes
#> 202 AMY1B;PRSS2;PRSS1;ATP1A2;PRSS3;CELA2A;CELA2B;CPA2;CPA1;CEL;CFTR;PNLIP;CELA3A;CELA3B;AMY2A;AMY2B;CPB1;PLA2G2D;PLA2G2A;CTRL;FXYD2;PNLIPRP2;PNLIPRP1;CTRB2;PLA2G10;CTRB1;PLA2G1B;ATP2A3;SLC4A4
#> 161 PDX1;BHLHA15;HNF1B;MAFA;NR5A2;IAPP;RFX6;FOXA2;ONECUT1;SLC2A2;INS;NKX2-2;NKX6-1;NEUROD1
#> 178 SST;AGTR2;PRSS2;VIPR2;PRSS1;PRSS3;AVPR1B;CCKBR;GRPR;PPY;EDN3;GABRA4;GABRB1;GRIK3;GCGR;GHRL;GLRA1;GLRA3;UCN3;NPY;CHRNB3;CHRNB2;GCG;IAPP;GHSR;GRIA2;CALCB;GRIA4;OPRM1;SSTR1;HTR1A;SSTR3;SSTR5;KNG1;PRLHR;GABRG1;ADCYAP1;P2RX2;P2RX1;CRH;CNR1
#> Rank Database
#> 202 1 KEGG
#> 161 2 KEGG
#> 178 3 KEGGMultiple gene set databases
You can also perform enrichment analysis against multiple gene set
databases at once. To specify multiple databases, you can pass a
vector of GMT file paths or a named
vector of GMT files to the EnrichIt
function. The names of the vector will be used as the database names in
the output. If you have pre-parsed GMT from the ParseGMT
function, you can pass a named list of parsed GMT files to the
EnrichIt function. The names of the list will be used as
the database names in the output.
# kegg_gmt <- system.file("extdata", "KEGG_2021_Human.gmt.gz", package = "enrichit")
# hallmark_gmt <- system.file("extdata", "MSigDB_Hallmark_2020.gmt.gz", package = "enrichit")
res <- EnrichIt(userlist, c(kegg_gmt, hallmark_gmt))
#> Warning in CheckUserList(userlist, db, use_matched_only = use_matched_only):
#> User list contains 335 unmatched genes. Showing first 10: A1CF, A4GNT, AADAC,
#> ADAMTS8, ADGRV1, AGR3, AJAP1, AKAIN1, ALKAL2, AMBP
#> Warning in CheckUserList(userlist, db, use_matched_only = use_matched_only):
#> User list contains 494 unmatched genes. Showing first 10: A1CF, A4GNT, AADAC,
#> ACMSD, ACSM2B, ACSM6, ACTL6B, ADAMTS8, ADGRV1, ADH1A
head(res[res$Database == "KEGG_2021_Human", ], n = 3)
#> Term Overlap P.value
#> 202 Pancreatic secretion 29/102 2.084613e-20
#> 161 Maturity onset diabetes of the young 14/26 3.925575e-15
#> 178 Neuroactive ligand-receptor interaction 41/341 8.073882e-14
#> Adjusted.P.value Odds.Ratio Combined.Score
#> 202 5.003072e-18 13.115022 594.3350
#> 161 4.710689e-13 37.671667 1249.6168
#> 178 6.459105e-12 4.552216 137.2382
#> Genes
#> 202 AMY1B;PRSS2;PRSS1;ATP1A2;PRSS3;CELA2A;CELA2B;CPA2;CPA1;CEL;CFTR;PNLIP;CELA3A;CELA3B;AMY2A;AMY2B;CPB1;PLA2G2D;PLA2G2A;CTRL;FXYD2;PNLIPRP2;PNLIPRP1;CTRB2;PLA2G10;CTRB1;PLA2G1B;ATP2A3;SLC4A4
#> 161 PDX1;BHLHA15;HNF1B;MAFA;NR5A2;IAPP;RFX6;FOXA2;ONECUT1;SLC2A2;INS;NKX2-2;NKX6-1;NEUROD1
#> 178 SST;AGTR2;PRSS2;VIPR2;PRSS1;PRSS3;AVPR1B;CCKBR;GRPR;PPY;EDN3;GABRA4;GABRB1;GRIK3;GCGR;GHRL;GLRA1;GLRA3;UCN3;NPY;CHRNB3;CHRNB2;GCG;IAPP;GHSR;GRIA2;CALCB;GRIA4;OPRM1;SSTR1;HTR1A;SSTR3;SSTR5;KNG1;PRLHR;GABRG1;ADCYAP1;P2RX2;P2RX1;CRH;CNR1
#> Rank Database
#> 202 1 KEGG_2021_Human
#> 161 2 KEGG_2021_Human
#> 178 3 KEGG_2021_Human
head(res[res$Database == "MSigDB_Hallmark_2020", ], n = 3)
#> Term Overlap P.value Adjusted.P.value Odds.Ratio
#> 501 Pancreas Beta Cells 19/40 9.856249e-19 3.252562e-17 29.446579
#> 49 KRAS Signaling Dn 20/200 3.799836e-06 6.269730e-05 3.592593
#> 451 Pperoxisome 7/104 4.112513e-02 4.523764e-01 2.293228
#> Combined.Score
#> 501 1220.884924
#> 49 44.837541
#> 451 7.318004
#> Genes
#> 501 CHGA;ABCC8;PDX1;GCG;G6PC2;SST;IAPP;INSM1;DCX;FOXA2;PCSK2;PCSK1;SLC2A2;INS;SCGN;NKX2-2;NKX6-1;PAK3;NEUROD1
#> 49 EGF;CNTFR;TENT5C;CLPS;TCL1A;IL12B;GP2;MYOT;SERPINA10;KCNQ2;ZBTB16;CAPN9;CPA2;CALCB;CPB1;SLC38A3;MYH7;NPHS1;ARHGDIG;NOS1
#> 451 RXRG;CEL;CACNA1B;ABCC8;ALB;SERPINA6;TTR
#> Rank Database
#> 501 1 MSigDB_Hallmark_2020
#> 49 2 MSigDB_Hallmark_2020
#> 451 3 MSigDB_Hallmark_2020
res <- EnrichIt(userlist, list(KEGG = ParseGMT(kegg_gmt), Hallmark = ParseGMT(hallmark_gmt)))
#> Warning in CheckUserList(userlist, db, use_matched_only = use_matched_only):
#> User list contains 335 unmatched genes. Showing first 10: A1CF, A4GNT, AADAC,
#> ADAMTS8, ADGRV1, AGR3, AJAP1, AKAIN1, ALKAL2, AMBP
#> Warning in CheckUserList(userlist, db, use_matched_only = use_matched_only):
#> User list contains 494 unmatched genes. Showing first 10: A1CF, A4GNT, AADAC,
#> ACMSD, ACSM2B, ACSM6, ACTL6B, ADAMTS8, ADGRV1, ADH1A
head(res[res$Database == "KEGG", ], n = 3)
#> Term Overlap P.value
#> 202 Pancreatic secretion 29/102 2.084613e-20
#> 161 Maturity onset diabetes of the young 14/26 3.925575e-15
#> 178 Neuroactive ligand-receptor interaction 41/341 8.073882e-14
#> Adjusted.P.value Odds.Ratio Combined.Score
#> 202 5.003072e-18 13.115022 594.3350
#> 161 4.710689e-13 37.671667 1249.6168
#> 178 6.459105e-12 4.552216 137.2382
#> Genes
#> 202 AMY1B;PRSS2;PRSS1;ATP1A2;PRSS3;CELA2A;CELA2B;CPA2;CPA1;CEL;CFTR;PNLIP;CELA3A;CELA3B;AMY2A;AMY2B;CPB1;PLA2G2D;PLA2G2A;CTRL;FXYD2;PNLIPRP2;PNLIPRP1;CTRB2;PLA2G10;CTRB1;PLA2G1B;ATP2A3;SLC4A4
#> 161 PDX1;BHLHA15;HNF1B;MAFA;NR5A2;IAPP;RFX6;FOXA2;ONECUT1;SLC2A2;INS;NKX2-2;NKX6-1;NEUROD1
#> 178 SST;AGTR2;PRSS2;VIPR2;PRSS1;PRSS3;AVPR1B;CCKBR;GRPR;PPY;EDN3;GABRA4;GABRB1;GRIK3;GCGR;GHRL;GLRA1;GLRA3;UCN3;NPY;CHRNB3;CHRNB2;GCG;IAPP;GHSR;GRIA2;CALCB;GRIA4;OPRM1;SSTR1;HTR1A;SSTR3;SSTR5;KNG1;PRLHR;GABRG1;ADCYAP1;P2RX2;P2RX1;CRH;CNR1
#> Rank Database
#> 202 1 KEGG
#> 161 2 KEGG
#> 178 3 KEGG
head(res[res$Database == "Hallmark", ], n = 3)
#> Term Overlap P.value Adjusted.P.value Odds.Ratio
#> 501 Pancreas Beta Cells 19/40 9.856249e-19 3.252562e-17 29.446579
#> 49 KRAS Signaling Dn 20/200 3.799836e-06 6.269730e-05 3.592593
#> 451 Pperoxisome 7/104 4.112513e-02 4.523764e-01 2.293228
#> Combined.Score
#> 501 1220.884924
#> 49 44.837541
#> 451 7.318004
#> Genes
#> 501 CHGA;ABCC8;PDX1;GCG;G6PC2;SST;IAPP;INSM1;DCX;FOXA2;PCSK2;PCSK1;SLC2A2;INS;SCGN;NKX2-2;NKX6-1;PAK3;NEUROD1
#> 49 EGF;CNTFR;TENT5C;CLPS;TCL1A;IL12B;GP2;MYOT;SERPINA10;KCNQ2;ZBTB16;CAPN9;CPA2;CALCB;CPB1;SLC38A3;MYH7;NPHS1;ARHGDIG;NOS1
#> 451 RXRG;CEL;CACNA1B;ABCC8;ALB;SERPINA6;TTR
#> Rank Database
#> 501 1 Hallmark
#> 49 2 Hallmark
#> 451 3 HallmarkGenerate clusterProfiler style result
The EnrichIt function can also generate results in the
clusterProfiler style. To do this, you can set the
style argument to "clusterProfiler".
head(EnrichIt(userlist, kegg_gmt, style = "clusterProfiler"), n = 3)
#> Warning in CheckUserList(userlist, db, use_matched_only = use_matched_only):
#> User list contains 335 unmatched genes. Showing first 10: A1CF, A4GNT, AADAC,
#> ADAMTS8, ADGRV1, AGR3, AJAP1, AKAIN1, ALKAL2, AMBP
#> ID Description GeneRatio
#> 202 KEGG_2021_Human_202 Pancreatic secretion 29/279
#> 161 KEGG_2021_Human_161 Maturity onset diabetes of the young 14/279
#> 178 KEGG_2021_Human_178 Neuroactive ligand-receptor interaction 41/279
#> BgRatio pvalue p.adjust qvalue
#> 202 102/8078 2.480401e-19 5.952961e-17 5.274115e-17
#> 161 26/8078 1.658281e-14 1.989937e-12 1.763014e-12
#> 178 341/8078 1.317274e-12 1.053819e-10 9.336465e-11
#> geneID
#> 202 AMY1B/PRSS2/PRSS1/ATP1A2/PRSS3/CELA2A/CELA2B/CPA2/CPA1/CEL/CFTR/PNLIP/CELA3A/CELA3B/AMY2A/AMY2B/CPB1/PLA2G2D/PLA2G2A/CTRL/FXYD2/PNLIPRP2/PNLIPRP1/CTRB2/PLA2G10/CTRB1/PLA2G1B/ATP2A3/SLC4A4
#> 161 PDX1/BHLHA15/HNF1B/MAFA/NR5A2/IAPP/RFX6/FOXA2/ONECUT1/SLC2A2/INS/NKX2-2/NKX6-1/NEUROD1
#> 178 SST/AGTR2/PRSS2/VIPR2/PRSS1/PRSS3/AVPR1B/CCKBR/GRPR/PPY/EDN3/GABRA4/GABRB1/GRIK3/GCGR/GHRL/GLRA1/GLRA3/UCN3/NPY/CHRNB3/CHRNB2/GCG/IAPP/GHSR/GRIA2/CALCB/GRIA4/OPRM1/SSTR1/HTR1A/SSTR3/SSTR5/KNG1/PRLHR/GABRG1/ADCYAP1/P2RX2/P2RX1/CRH/CNR1
#> Count Database
#> 202 29 KEGG_2021_Human
#> 161 14 KEGG_2021_Human
#> 178 41 KEGG_2021_HumanVisualization
The result of the EnrichIt function can be visualized
using the EnrichmentPlot function from the scplotter
package. The EnrichmentPlot function supports various plot
types, including bar plots, dot plots, network plots, word clouds,
etc.
res <- EnrichIt(userlist, c(kegg_gmt, hallmark_gmt), style = "clusterProfiler")
#> Warning in CheckUserList(userlist, db, use_matched_only = use_matched_only):
#> User list contains 335 unmatched genes. Showing first 10: A1CF, A4GNT, AADAC,
#> ADAMTS8, ADGRV1, AGR3, AJAP1, AKAIN1, ALKAL2, AMBP
#> Warning in CheckUserList(userlist, db, use_matched_only = use_matched_only):
#> User list contains 494 unmatched genes. Showing first 10: A1CF, A4GNT, AADAC,
#> ACMSD, ACSM2B, ACSM6, ACTL6B, ADAMTS8, ADGRV1, ADH1A
EnrichmentPlot(res, split_by = "Database")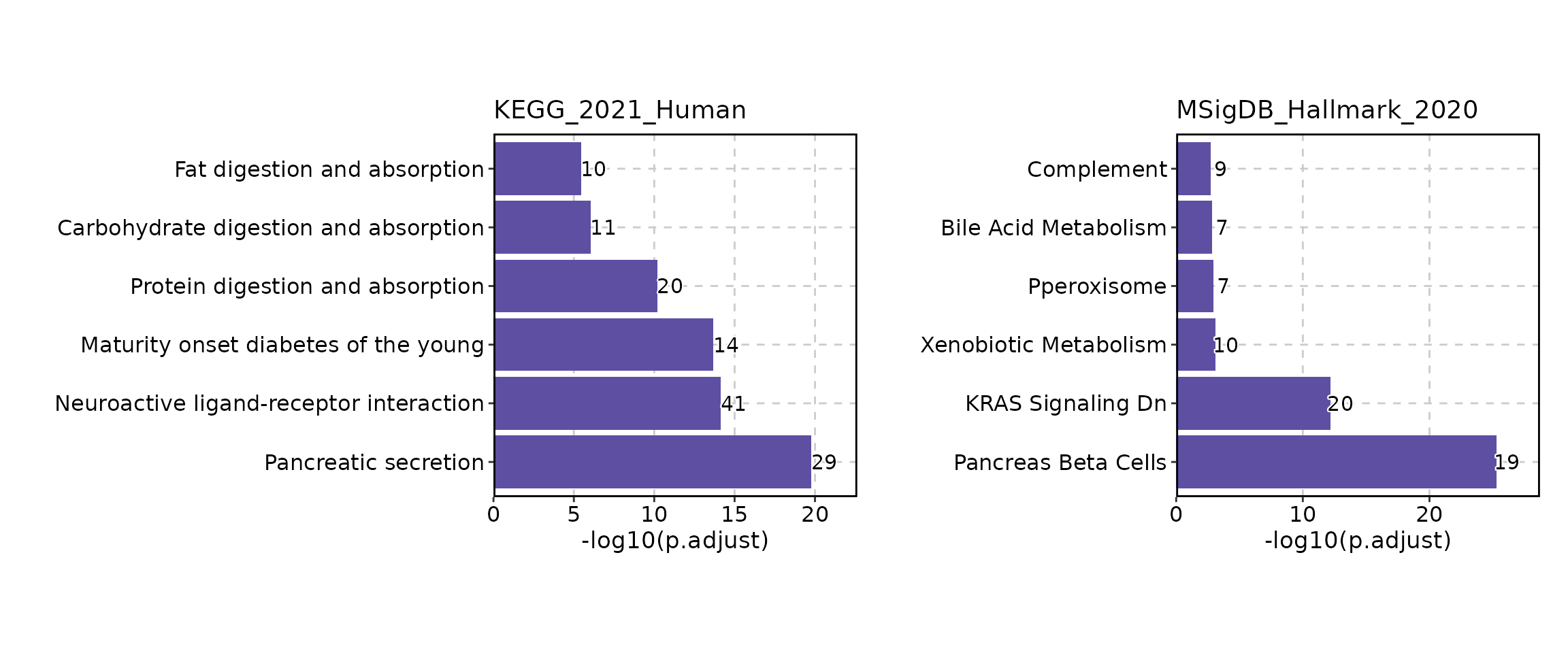
EnrichmentPlot(res, plot_type = "dot", split_by = "Database")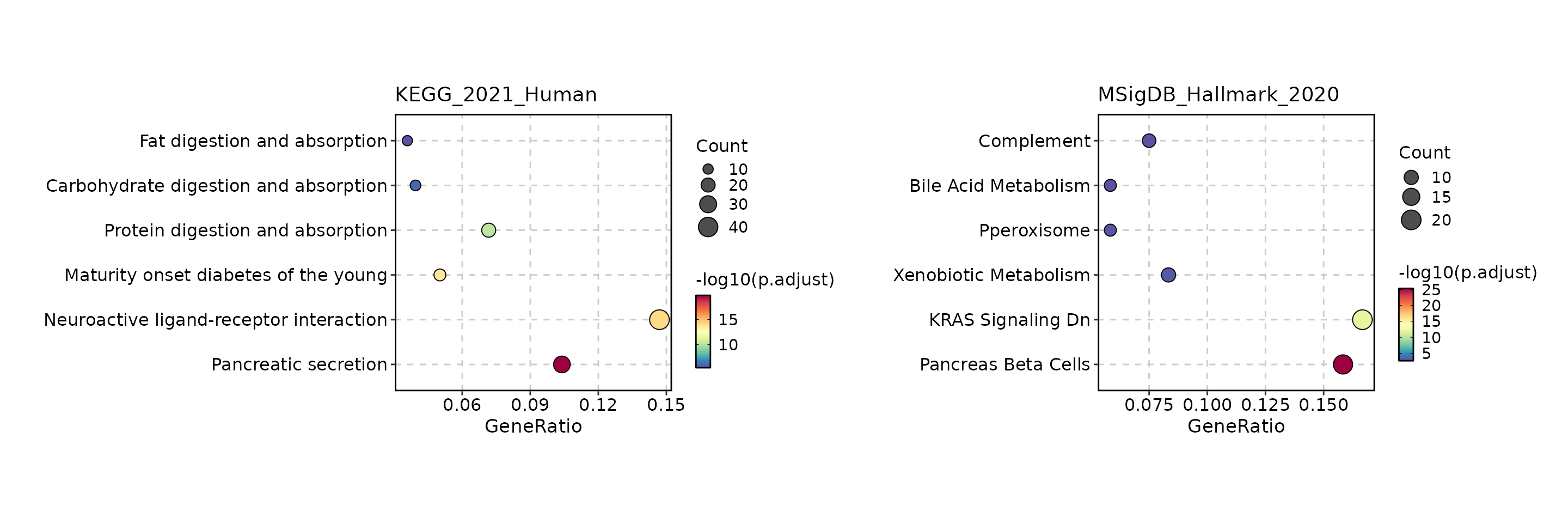
kegg_res <- res[res$Database == "KEGG_2021_Human", ]
EnrichmentPlot(kegg_res, plot_type = "network")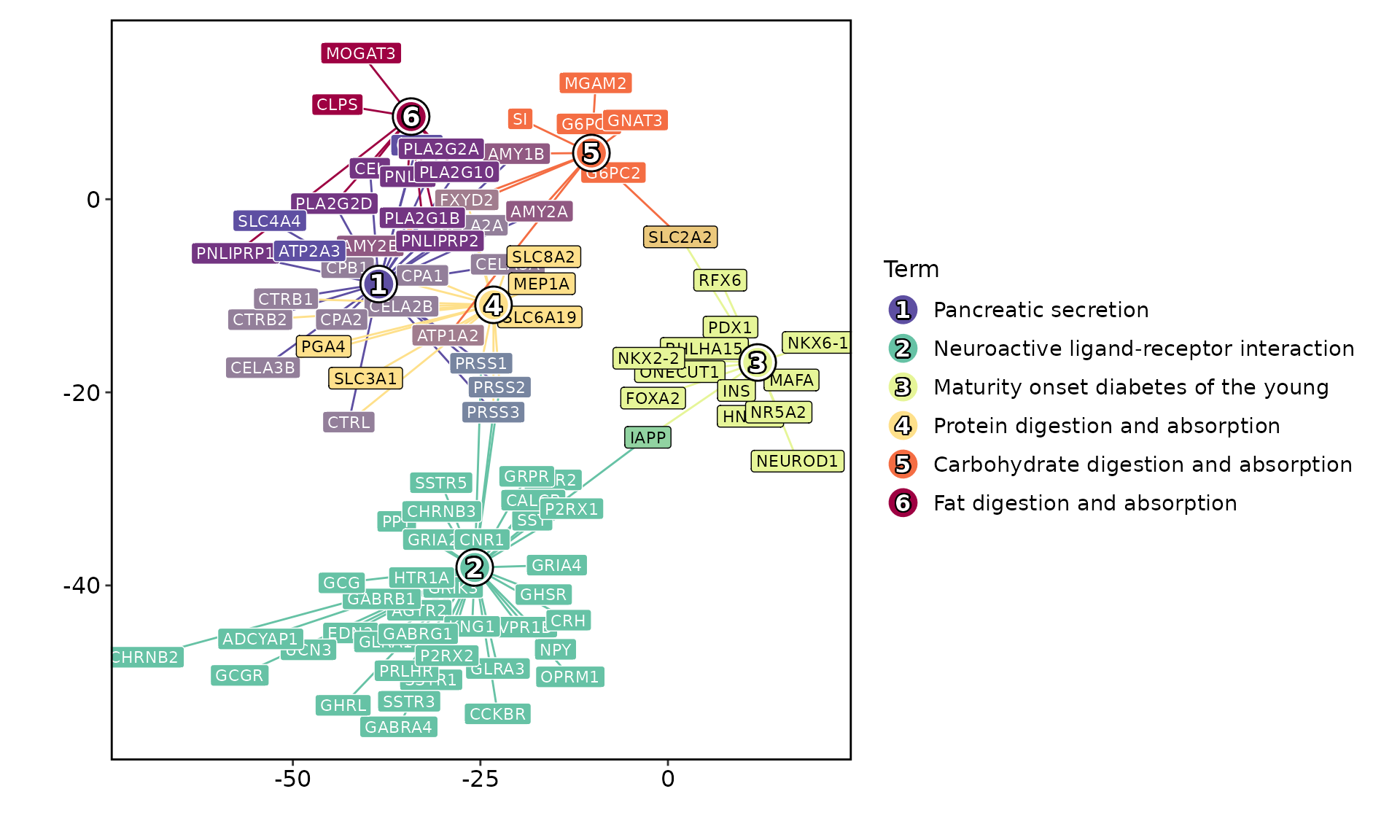
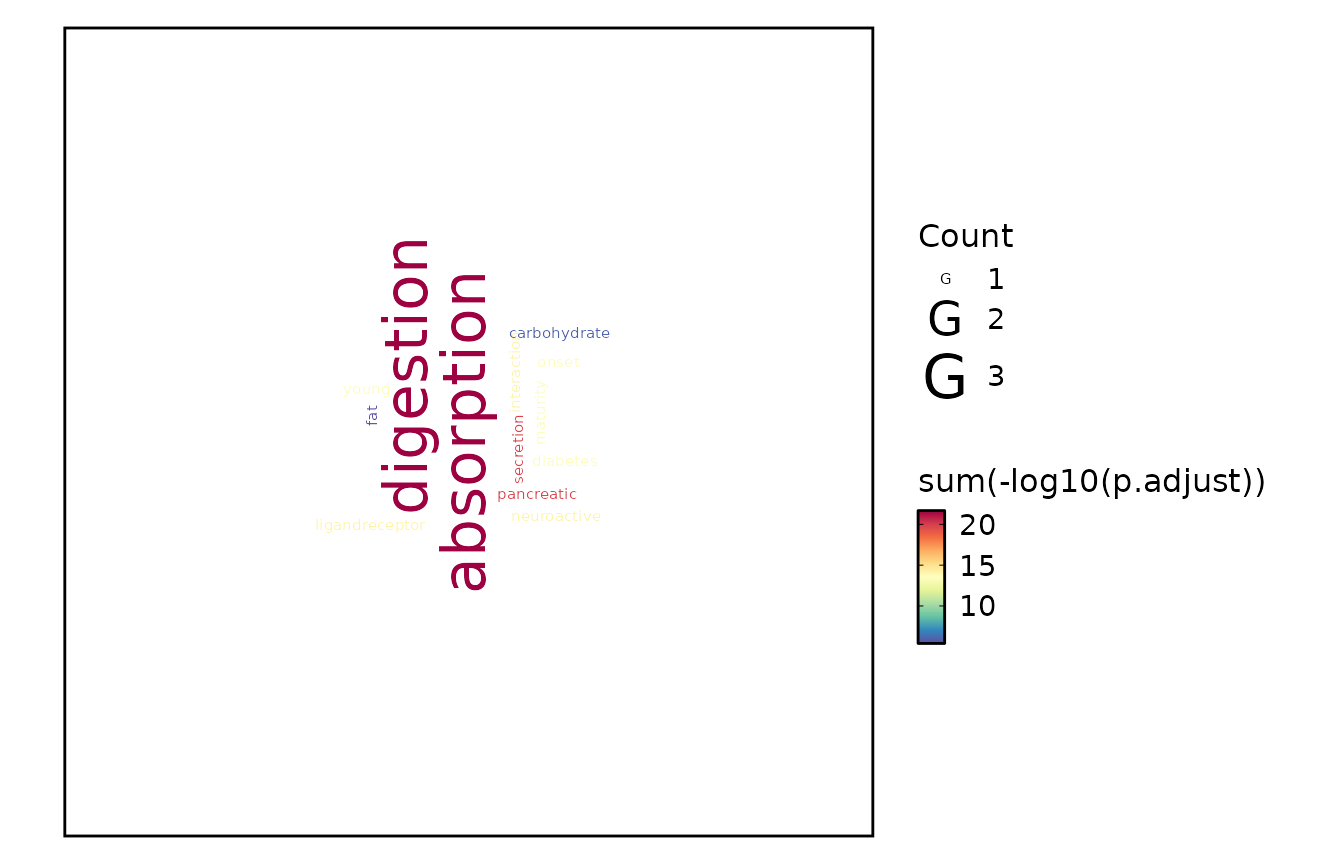
EnrichmentPlot(kegg_res, plot_type = "wordcloud")