Visualize differentially expressed genes
Usage
VizDEGs(
markers,
object = NULL,
plot_type = c("volcano", "volcano_log2fc", "volcano_pct", "jitter", "jitter_log2fc",
"jitter_pct", "heatmap_log2fc", "heatmap_pct", "dot_log2fc", "dot_pct", "heatmap",
"violin", "box", "bar", "ridge", "dot"),
subset_by = NULL,
subset_as_facet = FALSE,
comparison_by = NULL,
p_adjust = TRUE,
cutoff = NULL,
order_by = NULL,
select = ifelse(plot_type %in% c("volcano", "volcano_log2fc", "volcano_pct",
"jitter", "jitter_log2fc", "jitter_pct", "heatmap_log2fc", "heatmap_pct"), 5, 10),
outprefix = NULL,
devpars = list(res = 100),
more_formats = c(),
save_code = FALSE,
...
)Arguments
- markers
A data frame of markers, typically identified by
Seurat::FindMarkers()orSeurat::FindAllMarkers().- object
A Seurat object. Required for some plot types, see
plot_type.- plot_type
Type of plot to generate. Options include:
volcano/volcano_log2fc: Volcano plot with log2 fold change on x-axis and -log10(p-value) on y-axis. Ifp_adjustis TRUE, -log10(adjusted p-value) is used instead.volcano_pct: Volcano plot with difference in percentage of cells expressing the gene between two groups on x-axis and -log10(p-value) on y-axis. Ifp_adjustis TRUE, -log10(adjusted p-value) is used instead.jitter/jitter_log2fc: Jitter plot of log2 fold change for each gene. The x-axis is the groups defined bysubset_by, and the y-axis is log2 fold change. The size of the dots represents -log10(p-value) or -log10(adjusted p-value) ifp_adjustis TRUE.jitter_pct: Jitter plot of difference in percentage of cells expressing the gene between two groups for each gene. The x-axis is the groups defined bysubset_by, and the y-axis is the difference in percentage of cells expressing the gene between two groups. The size of the dots represents -log10(p-value) or -log10(adjusted p-value) ifp_adjustis TRUE.heatmap_log2fc: Heatmap of log2 fold change for each gene across groups defined bysubset_by. By specifyingcutoff, The heatmap cells will be labeled with "*" for p-value < cutoff, ifp_adjustis TRUE, adjusted p-value < cutoff.heatmap_pct: Heatmap of difference in percentage of cells expressing the gene between two groups for each gene across groups defined bysubset_by. By specifyingcutoff, The heatmap cells will be labeled with "*" for p-value < cutoff, ifp_adjustis TRUE, adjusted p-value < cutoff.dot_log2fc: Dot plot of log2 fold change for each gene across groups defined bysubset_by. The size of the dots represents -log10(p-value) or -log10(adjusted p-value) ifp_adjustis TRUE.dot_pct: Dot plot of difference in percentage of cells expressing the gene between two groups for each gene across groups defined bysubset_by. The size of the dots represents -log10(p-value) or -log10(adjusted p-value) ifp_adjustis TRUE.heatmap: Heatmap of expression values for each gene across groups defined bysubset_by. Requiresobject.violin: Violin plot of expression values for each gene across groups defined bysubset_by. Requiresobject.box: Box plot of expression values for each gene across groups defined bysubset_by. Requiresobject.bar: Bar plot of average expression values for each gene across groups defined bysubset_by. Requiresobject.ridge: Ridge plot of expression values for each gene across groups defined bysubset_by. Requiresobject.dot: Dot plot of expression values for each gene across groups defined bysubset_by. Requiresobject.
- subset_by
A column in markers indicating where the markers are identified from, e.g., cluster or condition. If object is provided, you can provide the corresponding metadata column in
subset_byto merge the markers with the object's metadata, using:as separator. For example, if the markers are identified from different clusters (identified byFindAllMarkers), and the object's ident column is "RNA_snn_res.0.8", you can setsubset_by = "cluster:RNA_snn_res.0.8". Note that for other columns to be merged, only the first value of each group defined bysubset_bywill be used. For some plots, this is used to split the markers into multiple plots, e.g., one plot for each cluster. Seeplot_typefor details.- subset_as_facet
Logical, whether to facet the plots by
subset_byif applicable.- comparison_by
The metadata column in markers indicating the comparion. When visualizing the expression values, this column should also be in the object's metadata. The values of this column should either a single value, indicating the comparison is between this group and all other cells (in the subset), Similar as
subset_by, you can provide the corresponding object metadata column by using:as separator. IfNULL, all markers are treated as from one comparison.- p_adjust
Logical, whether to use adjusted p-value for plots that involve p-values. Default is TRUE.
- cutoff
Numeric, p-value or adjusted p-value cutoff to label significance in heatmap plots. Default is NULL, no cutoff.
- order_by
A string of expression to order the markers within each group defined by
subset_by. In addition to the columns inmarkers, you can also use the columns from the object's metadata ifobjectis provided. The object's metadata will be merged withmarkersbysubset_by. Be carefull that only the first value of other columns will be used.- select
Number of top markers (ordered by
order_by) to select for each group defined bysubset_byor a string of expression to filter markers. It will be evaluated bydplyr::filter(). Forvolcano,volcano_log2fc,volcano_pct,jitter,jitter_log2fc, andjitter_pctplots, the selected markers will be labeled in the plot. FOr other plot types, only the selected markers will be plotted. Default is 5 forvolcano,volcano_log2fc,volcano_pct,jitter,jitter_log2fc,jitter_pct, and 10 for other plot types.- outprefix
Prefix of the output file
- devpars
List of parameters to save the plot
- more_formats
Additional formats to save the plot in addition to 'png'
- save_code
Whether to save the code to reproduce the plot
- ...
Arguments passed on to
scplotter::MarkersPlot
Examples
# \donttest{
degs <- RunSeuratDEAnalysis(scplotter::pancreas_sub, "SubCellType")
VizDEGs(degs, plot_type = "volcano_pct")
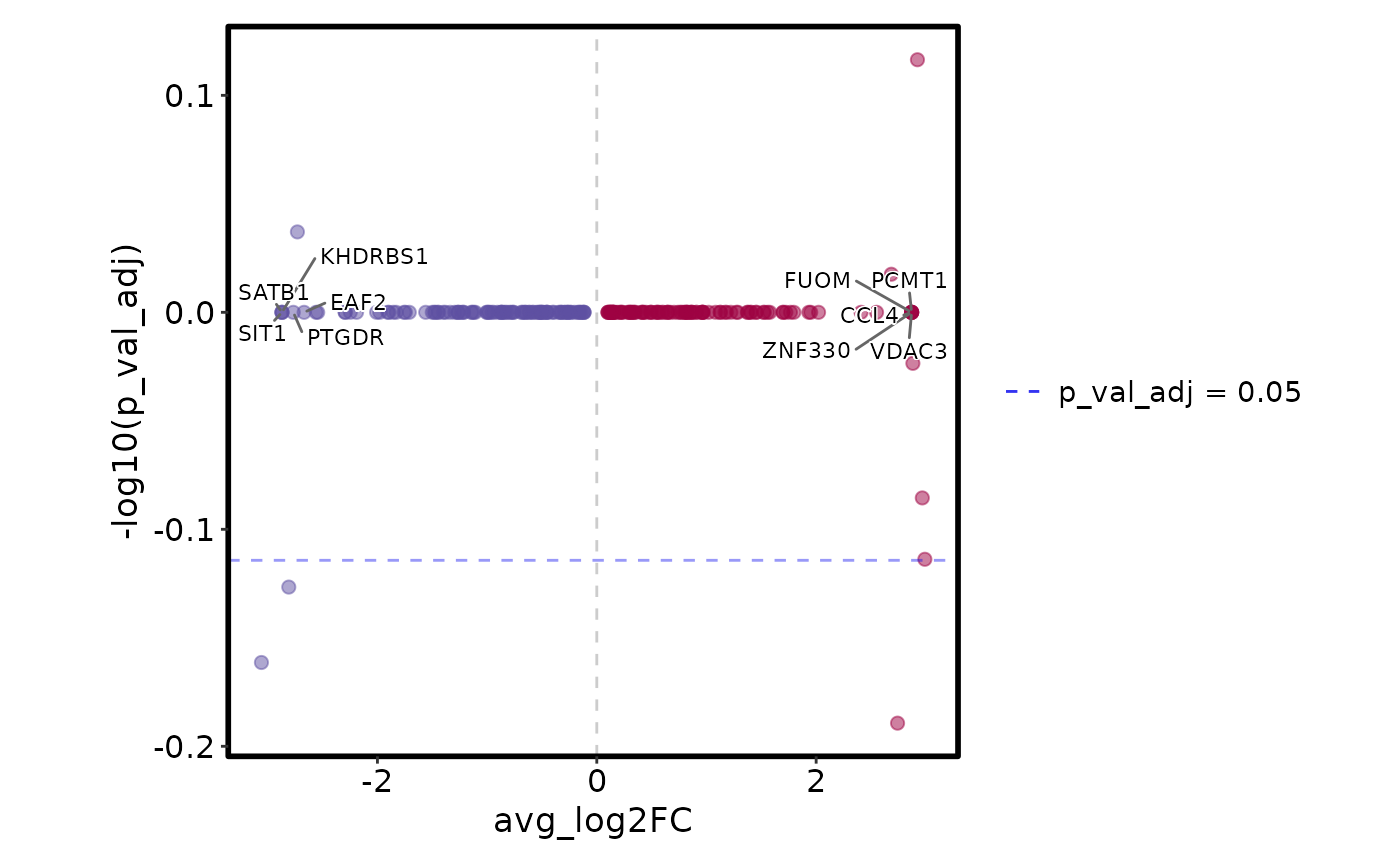
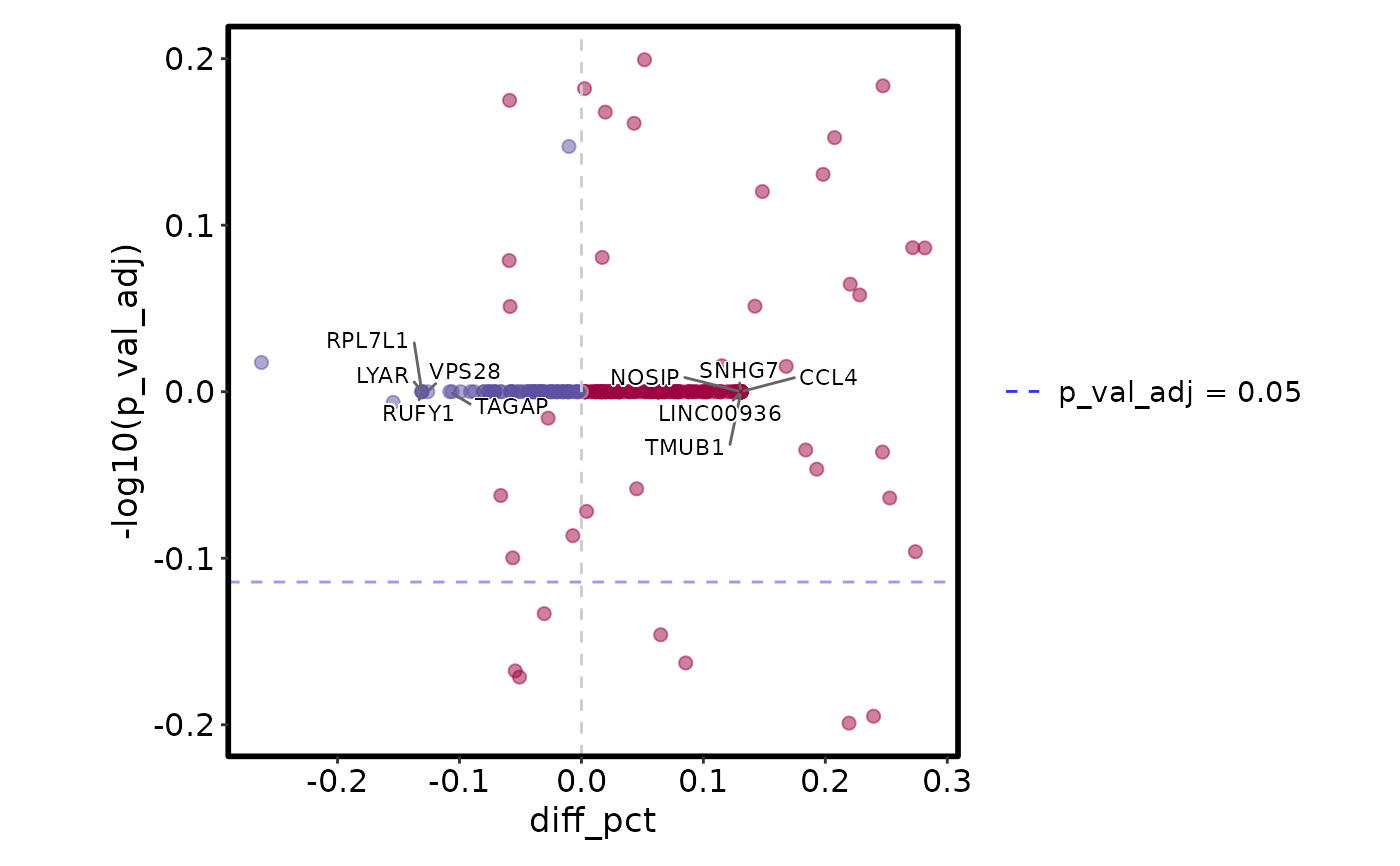 VizDEGs(degs, plot_type = "volcano_log2fc")
VizDEGs(degs, plot_type = "volcano_log2fc")
 VizDEGs(degs, plot_type = "jitter_log2fc", subset_by = "SubCellType")
VizDEGs(degs, plot_type = "jitter_log2fc", subset_by = "SubCellType")
 VizDEGs(degs,
plot_type = "heatmap_log2fc", cutoff = 0.05,
select = 5, subset_by = "SubCellType"
)
VizDEGs(degs,
plot_type = "heatmap_log2fc", cutoff = 0.05,
select = 5, subset_by = "SubCellType"
)
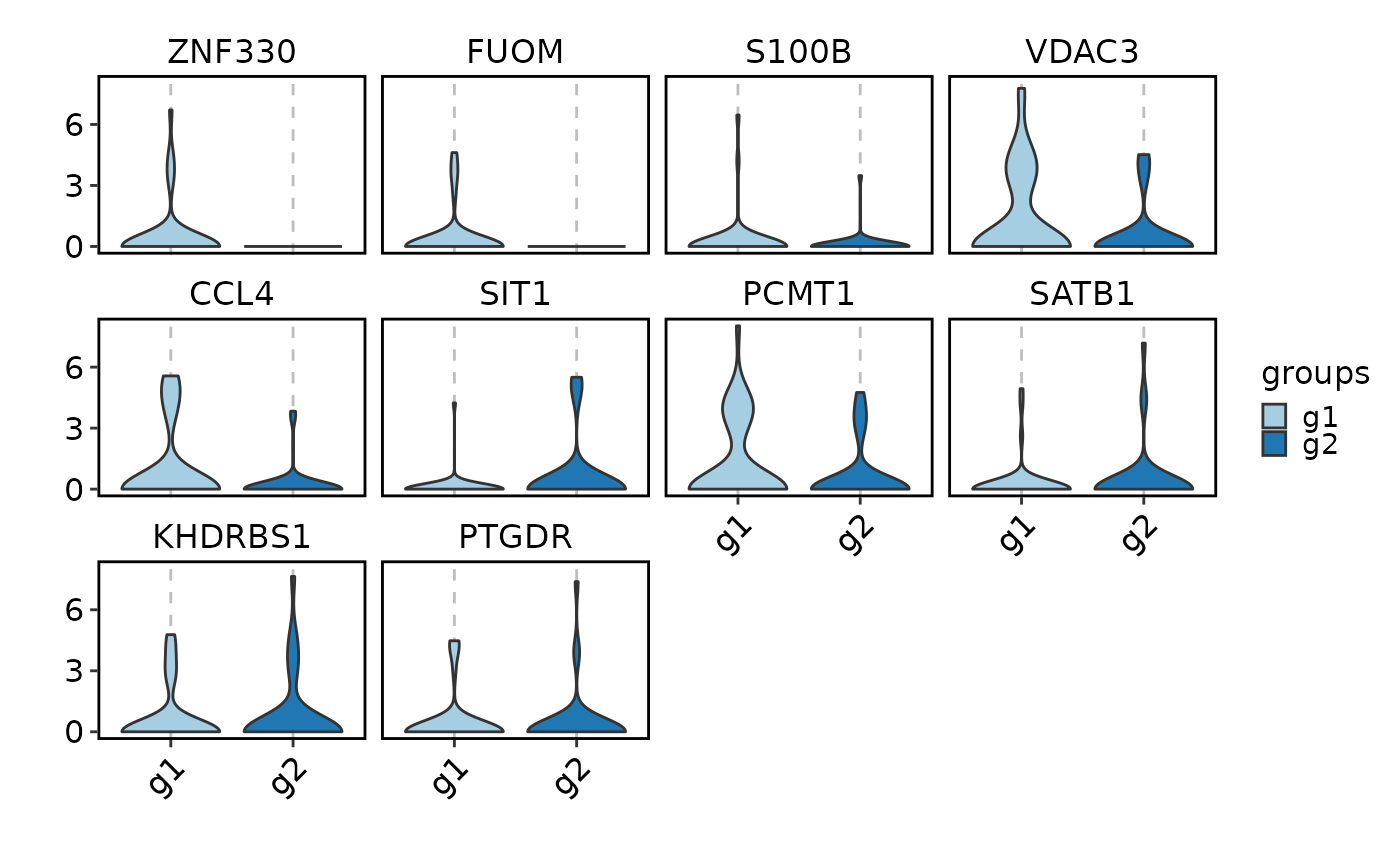
 # Visualize expression of the top DEGs
# Suppose we did comparison between G2M and S phase in each SubCellType
degs$Phase <- "G2M:S"
VizDEGs(degs,
object = scplotter::pancreas_sub, plot_type = "violin",
select = 2, comparison_by = "Phase", subset_by = "SubCellType"
)
#> Warning: Layer counts isn't present in the assay object; returning NULL
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
# Visualize expression of the top DEGs
# Suppose we did comparison between G2M and S phase in each SubCellType
degs$Phase <- "G2M:S"
VizDEGs(degs,
object = scplotter::pancreas_sub, plot_type = "violin",
select = 2, comparison_by = "Phase", subset_by = "SubCellType"
)
#> Warning: Layer counts isn't present in the assay object; returning NULL
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
#> Warning: Groups with fewer than two datapoints have been dropped.
#> ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
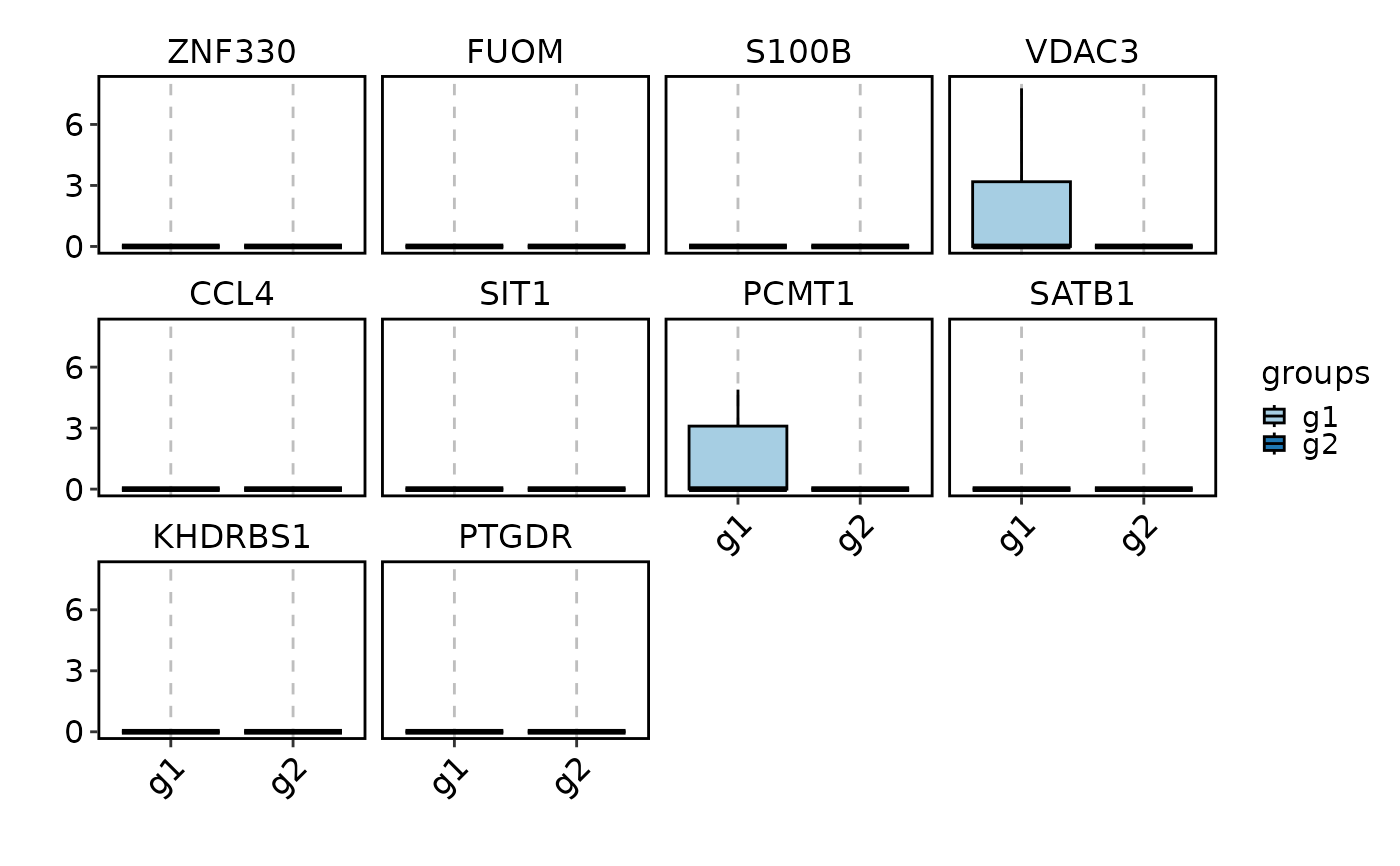
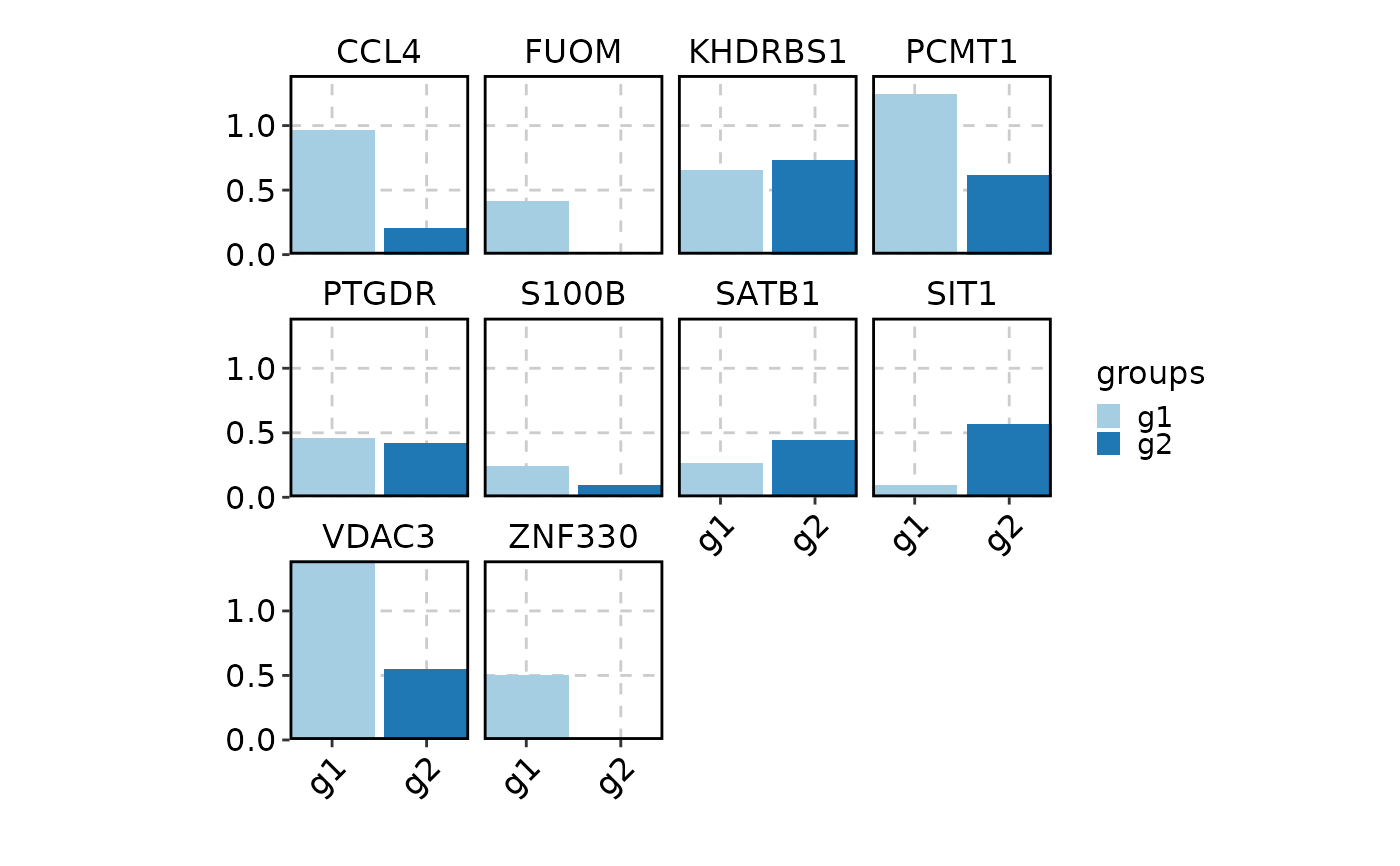 VizDEGs(degs,
object = scplotter::pancreas_sub, plot_type = "box",
select = 2, comparison_by = "Phase", subset_by = "SubCellType"
)
#> Warning: Layer counts isn't present in the assay object; returning NULL
VizDEGs(degs,
object = scplotter::pancreas_sub, plot_type = "box",
select = 2, comparison_by = "Phase", subset_by = "SubCellType"
)
#> Warning: Layer counts isn't present in the assay object; returning NULL
 VizDEGs(degs,
object = scplotter::pancreas_sub, plot_type = "bar",
select = 2, comparison_by = "Phase", subset_by = "SubCellType"
)
#> Warning: Layer counts isn't present in the assay object; returning NULL
VizDEGs(degs,
object = scplotter::pancreas_sub, plot_type = "bar",
select = 2, comparison_by = "Phase", subset_by = "SubCellType"
)
#> Warning: Layer counts isn't present in the assay object; returning NULL
 VizDEGs(degs,
object = scplotter::pancreas_sub, plot_type = "ridge",
select = 1, comparison_by = "Phase", subset_by = "SubCellType"
)
#> Warning: Layer counts isn't present in the assay object; returning NULL
#> Picking joint bandwidth of 0.283
#> Picking joint bandwidth of 0.252
#> Picking joint bandwidth of 0.196
#> Picking joint bandwidth of 0.0311
#> Picking joint bandwidth of 0.0338
#> Picking joint bandwidth of 0.382
#> Picking joint bandwidth of 0.0675
#> Picking joint bandwidth of 0.031
#> Picking joint bandwidth of 0.537
#> Picking joint bandwidth of 0.133
#> Picking joint bandwidth of 0.308
#> Picking joint bandwidth of 0.3
#> Picking joint bandwidth of 0.505
#> Picking joint bandwidth of 0.283
#> Picking joint bandwidth of 0.326
#> Picking joint bandwidth of 0.505
#> Picking joint bandwidth of 0.398
#> Picking joint bandwidth of 0.305
#> Picking joint bandwidth of 0.518
#> Picking joint bandwidth of 0.455
#> Picking joint bandwidth of 0.135
#> Picking joint bandwidth of 0.526
#> Picking joint bandwidth of 0.353
#> Picking joint bandwidth of 0.526
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Picking joint bandwidth of 0.283
#> Picking joint bandwidth of 0.252
#> Picking joint bandwidth of 0.196
#> Picking joint bandwidth of 0.0311
#> Picking joint bandwidth of 0.0338
#> Picking joint bandwidth of 0.382
#> Picking joint bandwidth of 0.0675
#> Picking joint bandwidth of 0.031
#> Picking joint bandwidth of 0.537
#> Picking joint bandwidth of 0.133
#> Picking joint bandwidth of 0.308
#> Picking joint bandwidth of 0.3
#> Picking joint bandwidth of 0.505
#> Picking joint bandwidth of 0.283
#> Picking joint bandwidth of 0.326
#> Picking joint bandwidth of 0.505
#> Picking joint bandwidth of 0.398
#> Picking joint bandwidth of 0.305
#> Picking joint bandwidth of 0.518
#> Picking joint bandwidth of 0.455
#> Picking joint bandwidth of 0.135
#> Picking joint bandwidth of 0.526
#> Picking joint bandwidth of 0.353
#> Picking joint bandwidth of 0.526
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
VizDEGs(degs,
object = scplotter::pancreas_sub, plot_type = "ridge",
select = 1, comparison_by = "Phase", subset_by = "SubCellType"
)
#> Warning: Layer counts isn't present in the assay object; returning NULL
#> Picking joint bandwidth of 0.283
#> Picking joint bandwidth of 0.252
#> Picking joint bandwidth of 0.196
#> Picking joint bandwidth of 0.0311
#> Picking joint bandwidth of 0.0338
#> Picking joint bandwidth of 0.382
#> Picking joint bandwidth of 0.0675
#> Picking joint bandwidth of 0.031
#> Picking joint bandwidth of 0.537
#> Picking joint bandwidth of 0.133
#> Picking joint bandwidth of 0.308
#> Picking joint bandwidth of 0.3
#> Picking joint bandwidth of 0.505
#> Picking joint bandwidth of 0.283
#> Picking joint bandwidth of 0.326
#> Picking joint bandwidth of 0.505
#> Picking joint bandwidth of 0.398
#> Picking joint bandwidth of 0.305
#> Picking joint bandwidth of 0.518
#> Picking joint bandwidth of 0.455
#> Picking joint bandwidth of 0.135
#> Picking joint bandwidth of 0.526
#> Picking joint bandwidth of 0.353
#> Picking joint bandwidth of 0.526
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Picking joint bandwidth of 0.283
#> Picking joint bandwidth of 0.252
#> Picking joint bandwidth of 0.196
#> Picking joint bandwidth of 0.0311
#> Picking joint bandwidth of 0.0338
#> Picking joint bandwidth of 0.382
#> Picking joint bandwidth of 0.0675
#> Picking joint bandwidth of 0.031
#> Picking joint bandwidth of 0.537
#> Picking joint bandwidth of 0.133
#> Picking joint bandwidth of 0.308
#> Picking joint bandwidth of 0.3
#> Picking joint bandwidth of 0.505
#> Picking joint bandwidth of 0.283
#> Picking joint bandwidth of 0.326
#> Picking joint bandwidth of 0.505
#> Picking joint bandwidth of 0.398
#> Picking joint bandwidth of 0.305
#> Picking joint bandwidth of 0.518
#> Picking joint bandwidth of 0.455
#> Picking joint bandwidth of 0.135
#> Picking joint bandwidth of 0.526
#> Picking joint bandwidth of 0.353
#> Picking joint bandwidth of 0.526
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Picking joint bandwidth of NaN
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
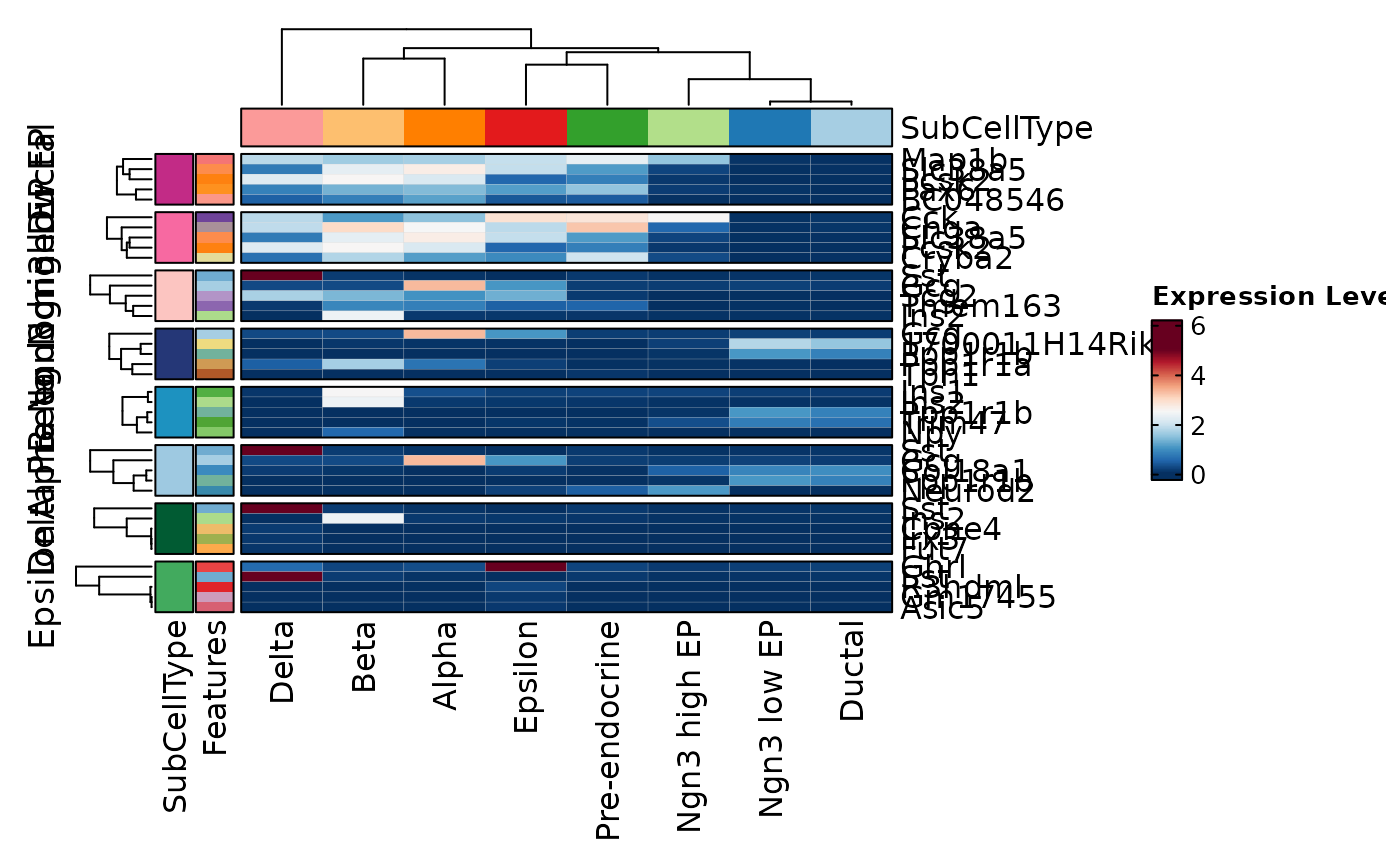
 VizDEGs(degs,
object = scplotter::pancreas_sub, plot_type = "heatmap",
cluster_columns = FALSE, comparison_by = "Phase", subset_by = "SubCellType"
)
#> Warning: Layer counts isn't present in the assay object; returning NULL
VizDEGs(degs,
object = scplotter::pancreas_sub, plot_type = "heatmap",
cluster_columns = FALSE, comparison_by = "Phase", subset_by = "SubCellType"
)
#> Warning: Layer counts isn't present in the assay object; returning NULL
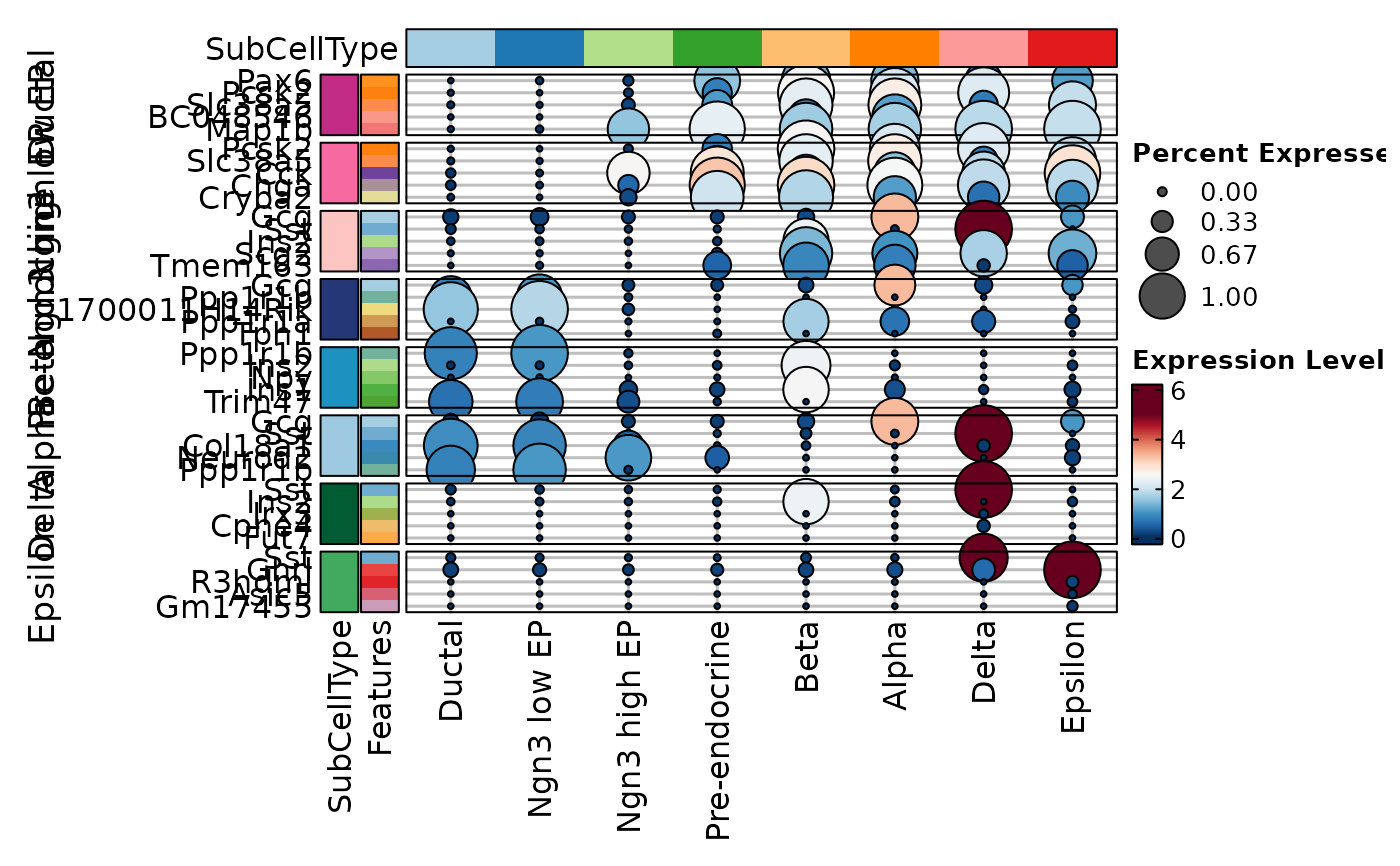
 VizDEGs(degs,
object = scplotter::pancreas_sub, plot_type = "dot",
select = 1, comparison_by = "Phase", subset_by = "SubCellType"
)
#> Warning: Layer counts isn't present in the assay object; returning NULL
VizDEGs(degs,
object = scplotter::pancreas_sub, plot_type = "dot",
select = 1, comparison_by = "Phase", subset_by = "SubCellType"
)
#> Warning: Layer counts isn't present in the assay object; returning NULL
 # }
# }