TCRClusterStats¶
Statistics of TCR clusters, generated by TCRClustering.
The statistics include
- The number of cells in each cluster (cluster size)
- Sample diversity using TCR clusters instead of TCR clones
- Shared TCR clusters between samples
Input¶
immfile: The immunarch object with TCR clusters attached
Output¶
outdir: Default:{{in.immfile | stem}}.tcrclusters_stats.
The output directory containing the stats and reports
Environment Variables¶
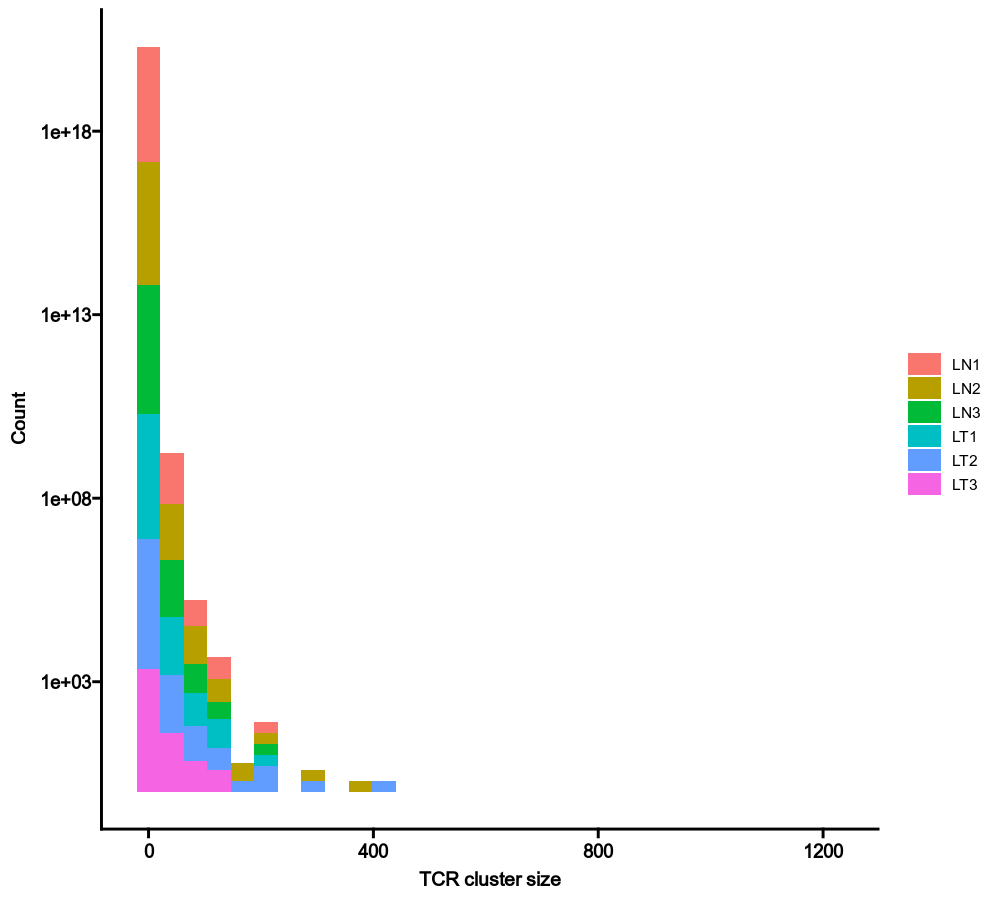
cluster_size(ns): The distribution of size of each cluster.by: Default:Sample.
The variables (column names) used to fill the histogram.
Only a single column is supported.devpars(ns): The parameters for the plotting device.width(type=int): Default:1000.
The width of the deviceheight(type=int): Default:900.
The height of the deviceres(type=int): Default:100.
The resolution of the device
cases(type=json): Default:{}.
If you have multiple cases, you can use this argument to specify them. The keys will be the names of the cases. The values will be passed to the corresponding arguments above. If any of these arguments are not specified, the values inenvs.cluster_sizewill be used. If NO cases are specified, the default case will be added, with the nameDEFAULT.
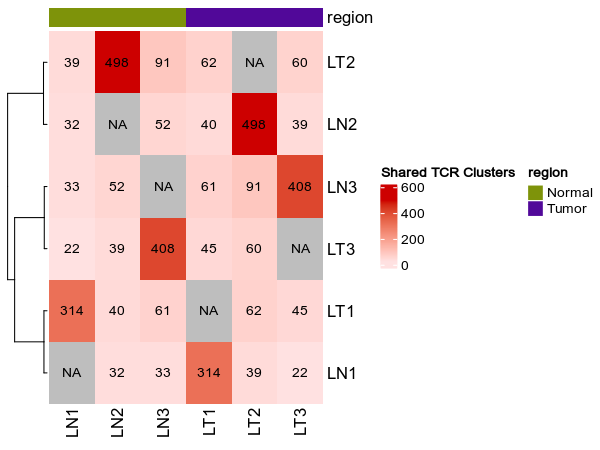
shared_clusters(ns): Stats about shared TCR clustersnumbers_on_heatmap(flag): Default:True.
Whether to show the numbers on the heatmap.heatmap_meta(list): Default:[].
The columns of metadata to show on the heatmap.cluster_rows(flag): Default:True.
Whether to cluster the rows on the heatmap.sample_order: The order of the samples on the heatmap.
Either a string separated by,or a list of sample names.
This only works for columns ifcluster_rowsisTrue.grouping: The groups to investigate the shared clusters.
If specified, venn diagrams will be drawn instead of heatmaps.
In such case,numbers_on_heatmapandheatmap_metawill be ignored.devpars(ns): The parameters for the plotting device.width(type=int): Default:1000.
The width of the deviceheight(type=int): Default:1000.
The height of the deviceres(type=int): Default:100.
The resolution of the device
cases(type=json): Default:{}.
If you have multiple cases, you can use this argument to specify them. The keys will be the names of the cases. The values will be passed to the corresponding arguments above. If any of these arguments are not specified, the values inenvs.shared_clusterswill be used. If NO cases are specified, the default case will be added, with the nameDEFAULT.
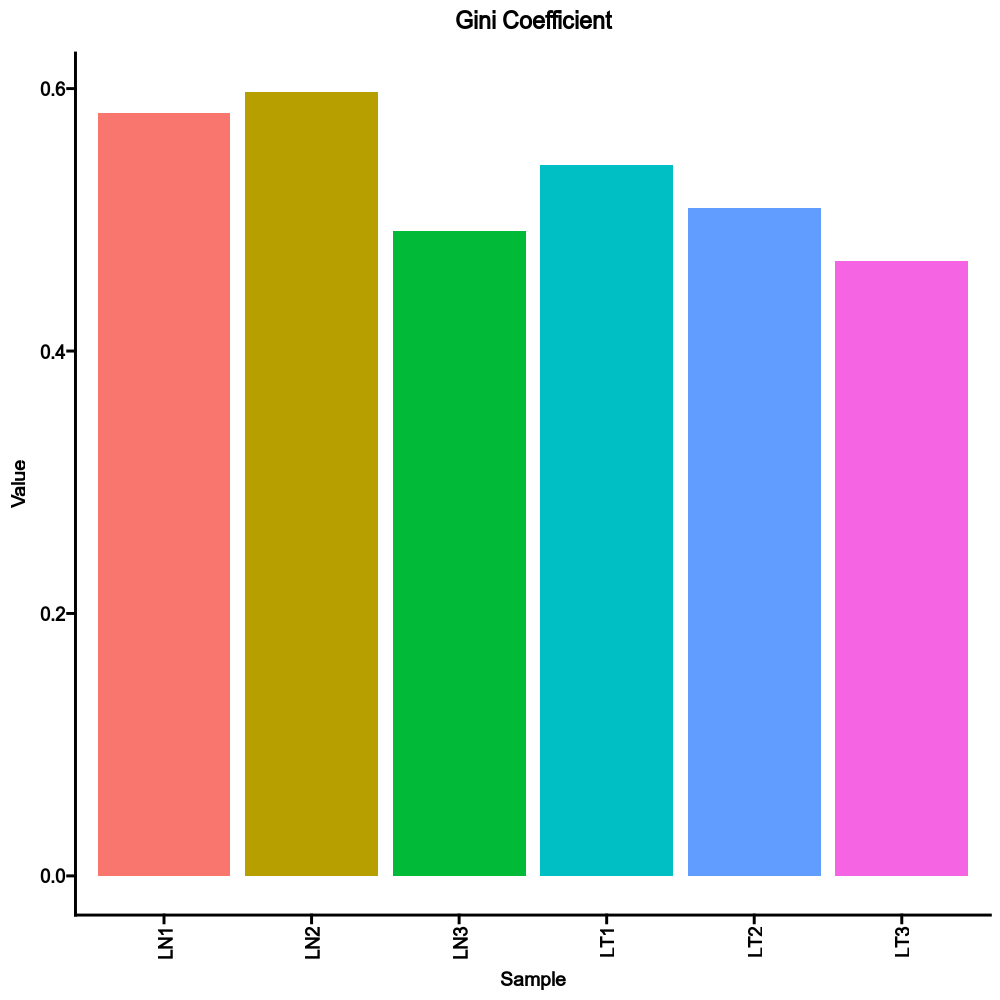
sample_diversity(ns): Sample diversity using TCR clusters instead of clones.by: The variables (column names) to group samples.
Multiple columns should be separated by,.method(choice): Default:gini.
The method to calculate diversity.gini: The Gini coefficient.
It measures the inequality among values of a frequency distribution (for example levels of income).gini.simp: The Gini-Simpson index.
It is the probability of interspecific encounter, i.e., probability that two entities represent different types.inv.simp: Inverse Simpson index.
It is the effective number of types that is obtained when the weighted arithmetic mean is used to quantify average proportional abundance of types in the dataset of interest.div: true diversity, or the effective number of types.
It refers to the number of equally abundant types needed for the average proportional abundance of the types to equal that observed in the dataset of interest where all types may not be equally abundant.
devpars(ns): The parameters for the plotting device.width(type=int): Default:1000.
The width of the deviceheight(type=int): Default:1000.
The height of the deviceres(type=int): Default:100.
The resolution of the device
cases(type=json): Default:{}.
If you have multiple cases, you can use this argument to specify them. The keys will be the names of the cases. The values will be passed to the corresponding arguments above. If any of these arguments are not specified, the values inenvs.sample_diversitywill be used. If NO cases are specified, the default case will be added, with the nameDEFAULT.
Examples¶
Cluster size¶
[TCRClusterStats.envs.cluster_size]
by = "Sample"
Shared clusters¶
[TCRClusterStats.envs.shared_clusters]
numbers_on_heatmap = true
heatmap_meta = ["region"]
Sample diversity¶
[TCRClusterStats.envs.sample_diversity]
method = "gini"
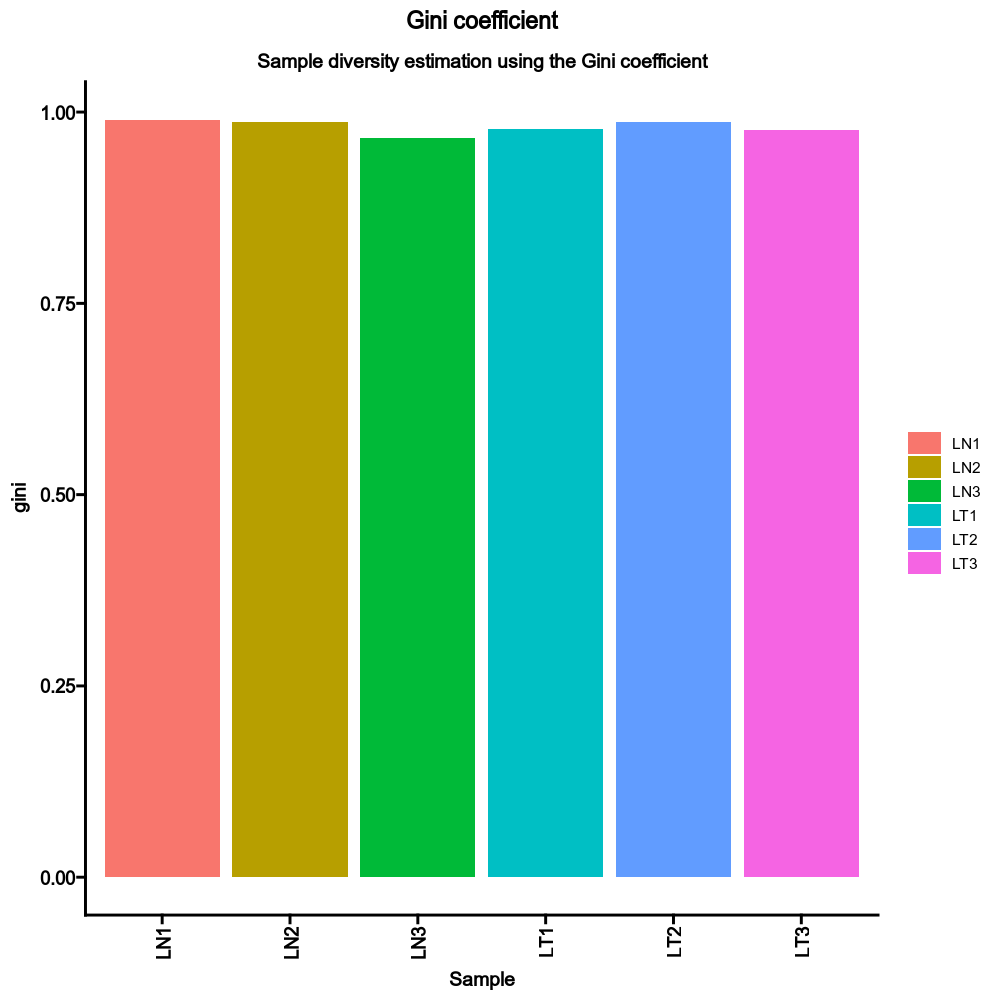
Compared to the sample diversity using TCR clones: